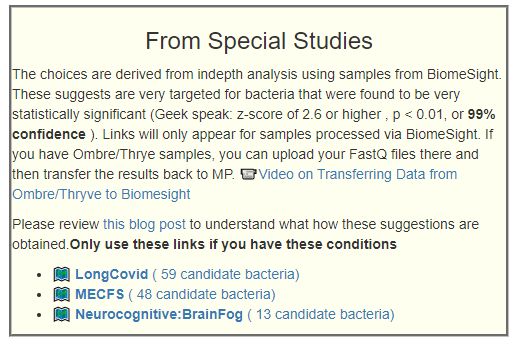
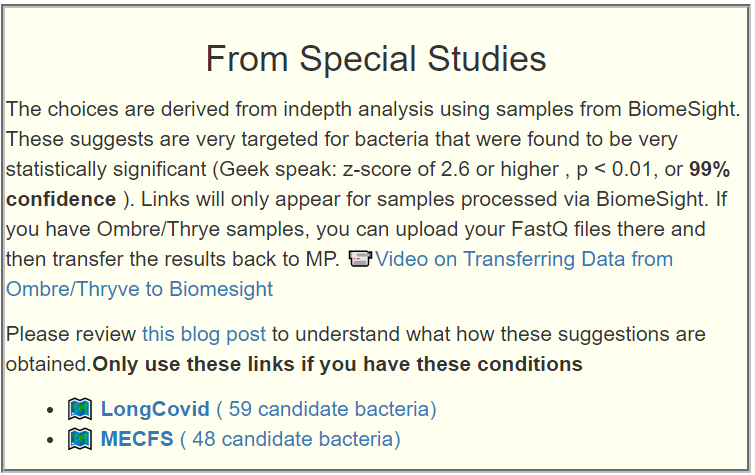
This is a common symptom for both ME/CFS and Long COVID. This is reported often in samples, and thus being examined if it reaches our threshold for inclusion as defined in A new specialized selection of suggestions links.
Beyond the goal of identifying bacteria involved, I am curious on the intersection of the bacteria with ME/CFS and Long COVID – i.e. bacteria in common and not in common.
Study Populations:
| Symptom | Reference | Study |
| Post-Exertional Malaise (PEM) | 1086 | 62 |
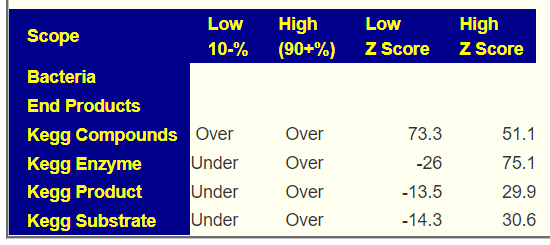
- Bacteria Detected with z-score > 2.6: found 181 items, highest value was 6.2
- Enzymes Detected with z-score > 2.6: found 237 items, highest value was 7.0
- Compound Detected with z-score > 2.6: found ZERO items
The highest z-scores above are less than other symptoms. There are two possible reasons:
- Smaller Study Population
- A more varied population in the study group.
Interesting Significant Bacteria
All bacteria found significant had too low levels.
We have two dominant bacteria group, both Bifidobacterium and Sporolactobacillus. The latter we know little about. I should point out that these bacteria may not be the cause, rather they may be ‘the canaries in the coal mine’ of the microbiome. These studies’ methodology determines association and not causality.
| Bacteria (Rank) | Reference Mean | Study Mean | z-score |
| Sporolactobacillus (genus) | 174 | 60 | 6.2 |
| Sporolactobacillus putidus (species) | 174 | 60 | 6.2 |
| Sporolactobacillaceae (family) | 173 | 60 | 6.2 |
| Bifidobacterium cuniculi (species) | 81 | 24 | 5.9 |
| Bifidobacterium asteroides (species) | 58 | 23 | 5.9 |
| [Ruminococcus] gnavus (species) | 7421 | 3336 | 5.4 |
| Mediterraneibacter (genus) | 7870 | 3717 | 5.3 |
Interesting Enzymes
All enzymes found significant had too low levels.
I will leave it to the reader to go to Kyoto Encyclopedia of Genes and Genomes to learn about these enzymes (a steep learning curve).
There are some items of special interest appearing which I drill into below.
| Enzyme | Z-Score |
| hydrogen-sulfide:ferredoxin oxidoreductase (1.8.7.1) | 7 |
| D-fructose:ubiquinone 5-oxidoreductase (1.1.5.14) | 6.2 |
| D-fructosyl-L-lysine 3-epimerase (5.1.3.41) | 6.1 |
| L-tryptophan carboxy-lyase (4.1.1.105) | 6 |
| aromatic-L-amino-acid carboxy-lyase (4.1.1.28) | 6 |
| CTP:N-acylneuraminate cytidylyltransferase (2.7.7.43) | 5.9 |
| protein-Npi-phospho-L-histidine:L-ascorbate Npi-phosphotransferase (2.7.1.194) | 5.8 |
| propane-1,2-diol hydro-lyase (propanal-forming) (4.2.1.28) | 5.7 |
| N-methylhydantoin amidohydrolase (ATP-hydrolysing) (3.5.2.14) | 5.5 |
| D-ribopyranose furanomutase (5.4.99.62) | 5.5 |
| 3-dehydro-L-gulonate:NAD(P)+ 2-oxidoreductase (1.1.1.130) | 5.5 |
hydrogen-sulfide:ferredoxin oxidoreductase (1.8.7.1): This is connected to iron. The blood uses iron to carry oxygen, and thus an absence/low level could [speculation] result in an impact on the blood’s ability to deliver oxygen (thus fatigue).
D-fructose:ubiquinone 5-oxidoreductase (1.1.5.14): This is also connected to iron.
For those wishing to explore more, you may wish to read Oxidoreductase
It does hint at an experiment to try: After exercise, try a dosage of Ubiquinol to see if it influences things.
Common Bacteria Shifts Observed in ME/CFS
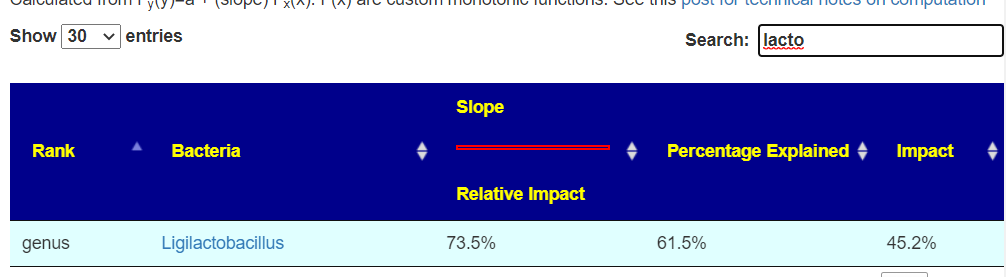
We have 45 bacteria in common, they are listed below. A LOT of them are bifidobacterium, and no lactobacillus. This implies that bifidobacterium probiotics may be a good choice for ME/CFS with PEM
| Tax_Name | Tax_rank |
| Thiorhodococcus pfennigii | species |
| Candidatus Tammella caduceiae | species |
| Veillonella atypica | species |
| Tammella | genus |
| Myxococcales | order |
| Gemella cuniculi | species |
| Bifidobacterium catenulatum | species |
| Nannocystineae | suborder |
| Olivibacter | genus |
| Campylobacterales | order |
| Epsilonproteobacteria | class |
| Campylobacteraceae | family |
| Pedobacter kwangyangensis | species |
| Haemophilus parainfluenzae | species |
| Haemophilus | genus |
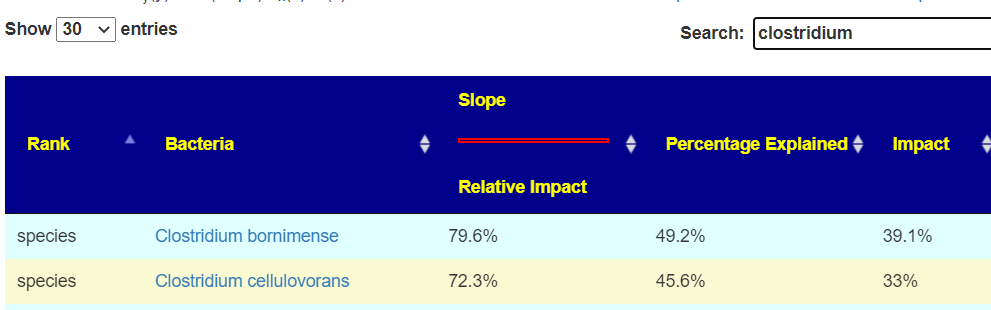
| Clostridium aestuarii | species |
| Sterolibacteriaceae | family |
| Lactococcus fujiensis | species |
| Bifidobacterium bifidum | species |
| Atopobium | genus |
| Balneola | genus |
| Balneola vulgaris | species |
| Balneolaceae | family |
| Thiobacillus | genus |
| Pigmentiphaga | genus |
| Thiobacillaceae | family |
| Balneolia | class |
| Balneolales | order |
| Balneolaeota | phylum |
| Ruminococcus flavefaciens | species |
| Hydrogenophilalia | class |
| Hydrogenophilales | order |
| Hydrogenophilaceae | family |
| Atopobiaceae | family |
| Veillonella dispar | species |
| Veillonella | genus |
| Clostridium chartatabidum | species |
| Actinobacillus pleuropneumoniae | species |
| Sporolactobacillaceae | family |
| Sporolactobacillus putidus | species |
| Sporolactobacillus | genus |
| Bifidobacterium kashiwanohense PV20-2 | strain |
| Bifidobacterium catenulatum subsp. kashiwanohense | subspecies |
| Bifidobacterium gallicum | species |
| Bifidobacterium cuniculi | species |
Common Bacteria Shifts Observed in Long COVID
We have 42 bacteria in common, they are listed below. We notice some interesting difference from above:
- Lactobacillus at the genus level as well as the retail probiotic Lactiplantibacillus plantarum (AKA Lactobacillus plantarum)
- Bifidobacterium is still there, but one of them is available as a retail probiotics.
- Bifidobacterium animalis
| Tax_Name | Tax_rank |
| Paenibacillus | genus |
| Veillonella | genus |
| Actinomycetaceae | family |
| Lactiplantibacillus plantarum | species |
| Actinomyces | genus |
| Flammeovirga | genus |
| Flammeovirga pacifica | species |
| Flammeovirgaceae | family |
| Lactiplantibacillus | genus |
| Phocaeicola massiliensis | species |
| Prosthecobacter | genus |
| Fusobacterium gonidiaformans | species |
| Candidatus Tammella caduceiae | species |
| Gammaproteobacteria | class |
| Tammella | genus |
| Coriobacteriaceae | family |
| Fusobacteria | phylum |
| Fusobacteriia | class |
| Fusobacteriales | order |
| Coriobacteriales | order |
| Bifidobacterium thermophilum | species |
| Dolichospermum curvum | species |
| Blautia wexlerae | species |
| Atopobiaceae | family |
| Actinobacillus pleuropneumoniae | species |
| Eggerthella lenta | species |
| Fusobacteriaceae | family |
| Atopobium | genus |
| Schaalia | genus |
| Bifidobacterium gallicum | species |
| Eggerthella | genus |
| Bifidobacterium animalis | species |
| Aerococcaceae | family |
| Coriobacteriia | class |
| Bifidobacterium cuniculi | species |
| Schaalia naturae | species |
| Phocaeicola sartorii | species |
| Leptospira licerasiae | species |
| Leptospiraceae | family |
| Leptospira | genus |
| Leptospirales | order |
| Alkalibacterium | genus |
Bottom Line
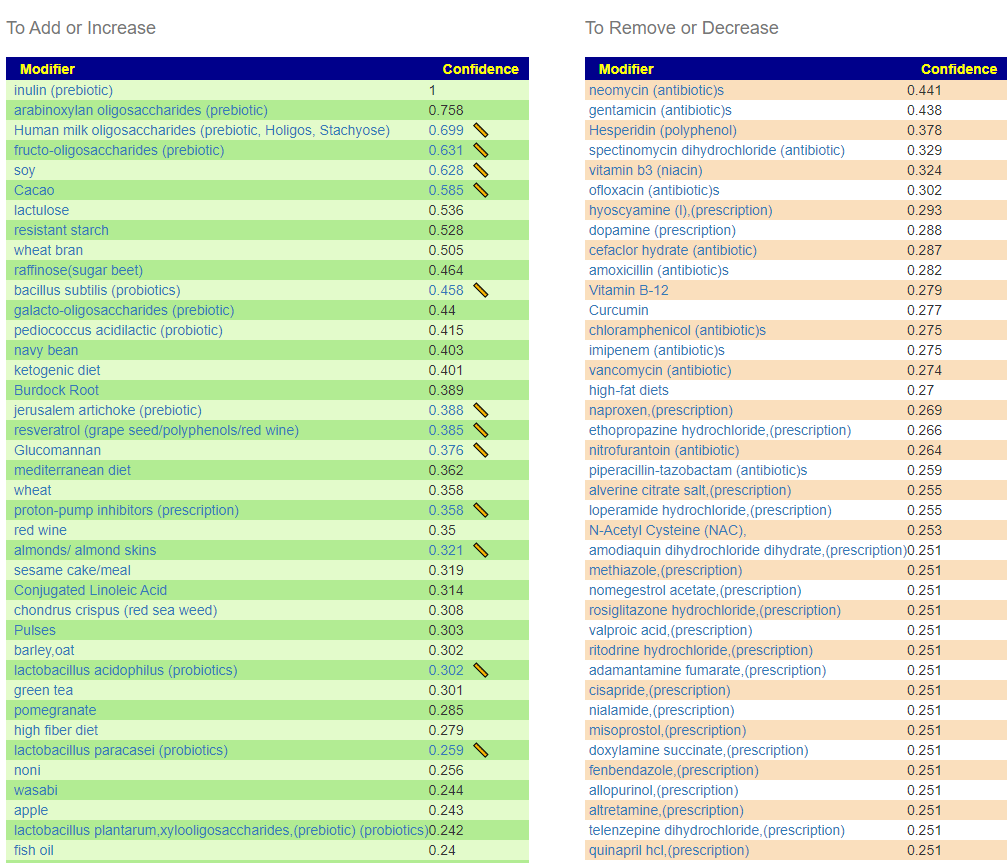
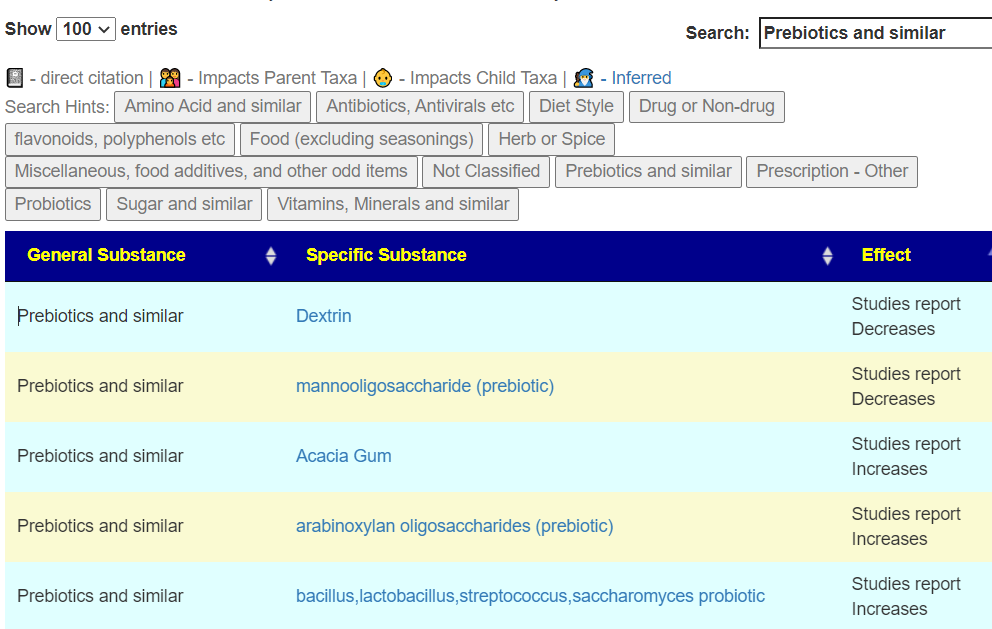
There appear to be differences between ME/CFS with PEM and Long COVID with PEM. The main difference is with Long COVID: Lactobacillus probiotics is a suggestion; for ME/CFS it is not.
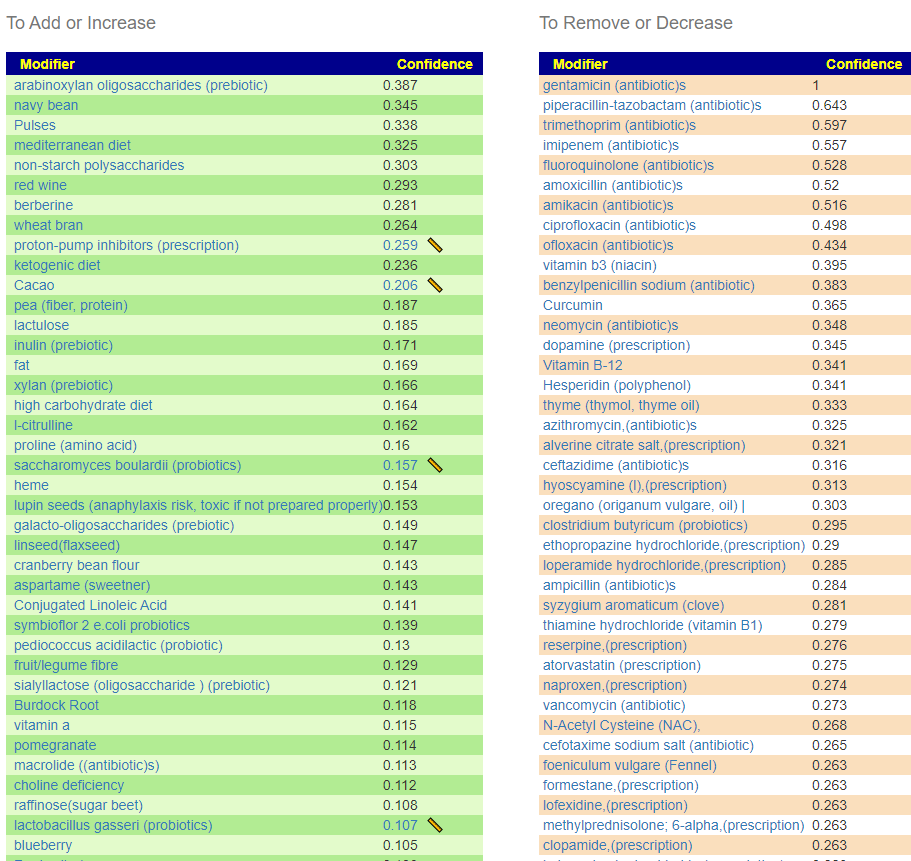
Remember suggestions that are specific to your unique microbiome are available on the Microbiome Prescription web site.





Recent Comments