Pages to assist with becoming a provider
- Home page for providers
- To test data transfers click here
- To help find NCBI Taxon numbers click here
Updated 2024 Guidance
The simplest ideal situation is make your lab results available as a CSV file, ideally with the following headers:
taxon no, percentage, percentile, other data you care to send
- Taxon number is from NCBI, search here
- Percentage is the percentage in the sample
- Percentile is against all other samples that you have processed. Percentile is very important
Most of our analysis is based on percentile ranking (which allows us to bypass issues with different lab pipelines). The same sample processed through different pipelines can give very different percentages but the percentile rankings are similar.
We can support (at no cost) the sending of the same data by JSON. We will create an API end-point for you.
Directly sending the data as JSON
This should only be done by their client (owner of the microbiome sample) from the provider site. This simplifies the transfer (more client friendly) and usually results in good will for the provider (as well as repeat business). Many people have switched to supported providers for subsequent tests because they wish to use the features on the Microbiome Prescription site.
The simplest format is a very very simple upload (post) a text file or JSON consisting of nothing more than:
Line 1: Email Address of user
Line 2-N: NCBI Taxon Number, Percentage, Percentile
That is it!!! We have a test page available for people who wish to try it (data is not saved).
When you wish to enable it so that it is saved, we will provide a key to you so that we know where the data is coming from. THERE IS NO COST. See statistics on uploads here.
This also clarifies to users whether they should or could be seeing a specific species, genus or family on their report. Below is a sample of the microbiome tree that is presented to the users. Any missing rank is estimated from it’s children.
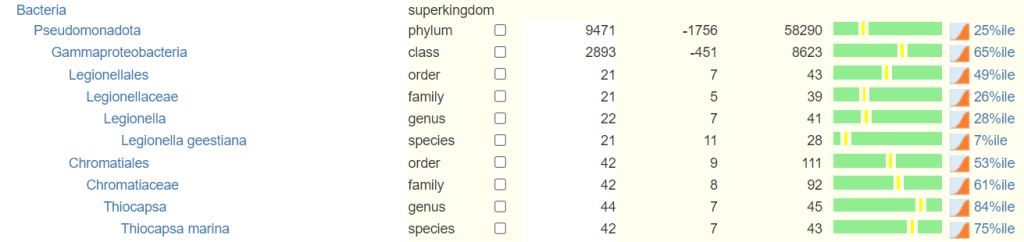
If percentile is not given, we estimate it from percentage by using data across all samples. This is unreliable and not recommended.
Route 1: Giving the client a file to download and then upload
Why NCBI Numbers?

The reasons are simple:
- various bacteria have dozens of names — on occasion, the name was deprecated and assigned to a different bacteria. It assures us that we have the right bacteria.
- We use the KEGG: Kyoto Encyclopedia of Genes and Genomes, and their data is all keyed to NCBI Taxon numbers
- It allows data to be stored in a more compact fashion (up to 60x smaller database) and allows faster processing of data (saving operating costs).
Please note that there are open source tools available to assist with finding the correct NCBI numbers, see https://youtu.be/VMi0dOeNQFA and https://youtu.be/B0zOSF8f0mo for an illustration.
Alternative: Use the NCBI Names
We have 3,635,527 names from NCBI in our database and can do a lookup of NCBI numbers by name. This is less precise — however, some providers use this route with over 99% of their names matching.
We request all taxonomic ranks to be specified
Our experience is that percentages become distorted (under reported) when we rollup from species to genus to family etc. Typically, there will be some undetermined species in each genus, family, etc. These will often be referred to as “unclassified Lactobacillus” etc.
The file will show the NAME for each of the taxon, so you can verify that the numbers are correct.
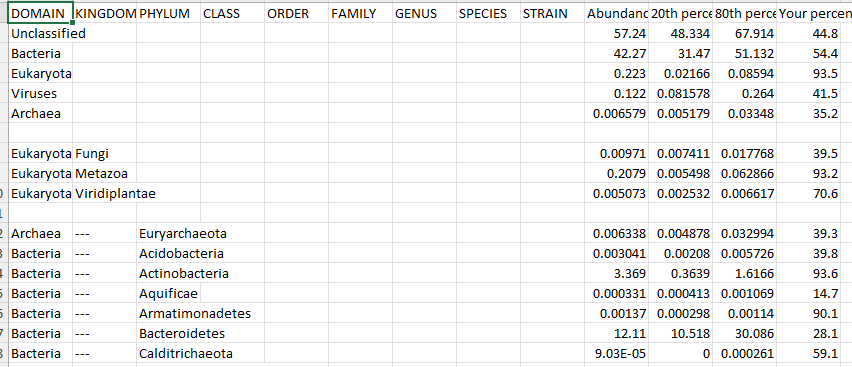
Below is an example from CosmosID, No taxon number is given BUT the names used match NCBI names so we can 100% name match.
Provider Motivations NOT to support
The following has become apparent from dialogs with some providers:
- They are not willing to spend money to get a suitable file created
- They do not have programming staff to code, i.e. they will need to hire a developer.
- They do not want to have their advice question by allowing second or third opinions of what people should take.
- Often they perceive this as generating support calls (paying more staff) dealing with customers’ questions
- Their business model depend on up-selling probiotics (huge profit margins), supplements, or meal plan. Customers getting second or third opinions risk lost of these profits
- Pure Ego issues.
Recent Comments