We have the Enzymes produces by a wide variety of strains on the KEGG: Kyoto Encyclopedia of Genes and Genomes. We can aggregate(i.e. average) this data up to the species level and then estimate the enzymes that a probiotic species or retail probiotic mixture may produce.
The next step is to identify the enzymes that a person is deficient in. I use the patent pending Kaltoft-Moltrup (KM) method to determine the bottom boundaries applying to percentile values of a significant population.
Some visual examples, with this point being around the 8th percentile.
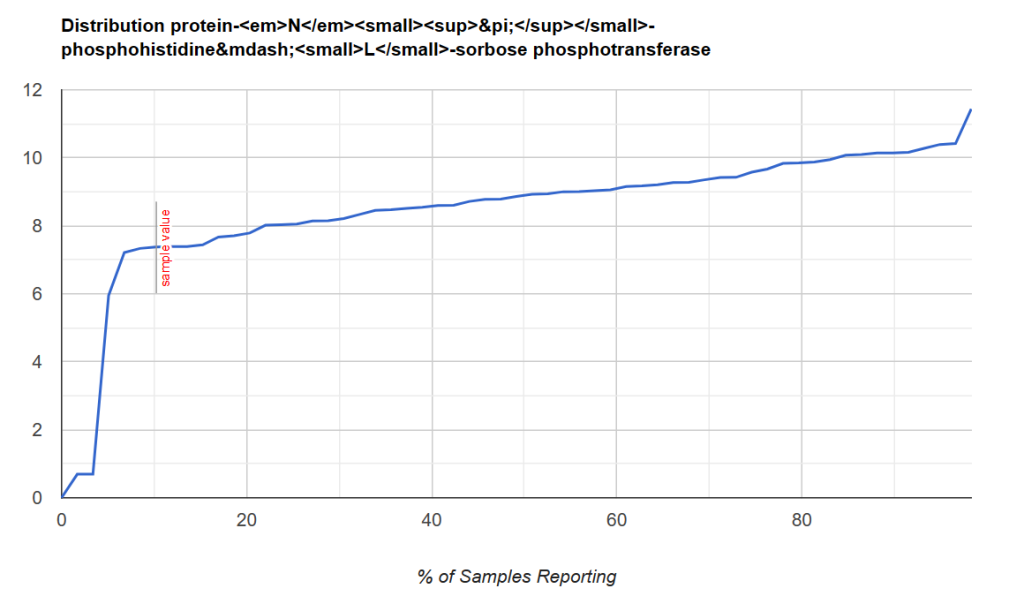
Another example with this point being between 2 and 7%ile
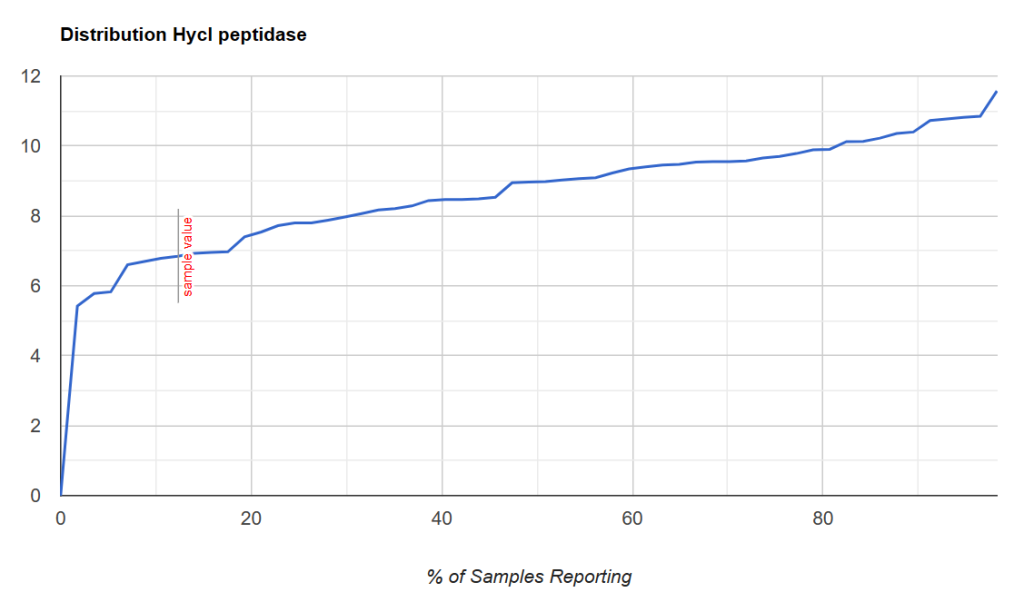
A sample with the cutoff being close to 20%ile.
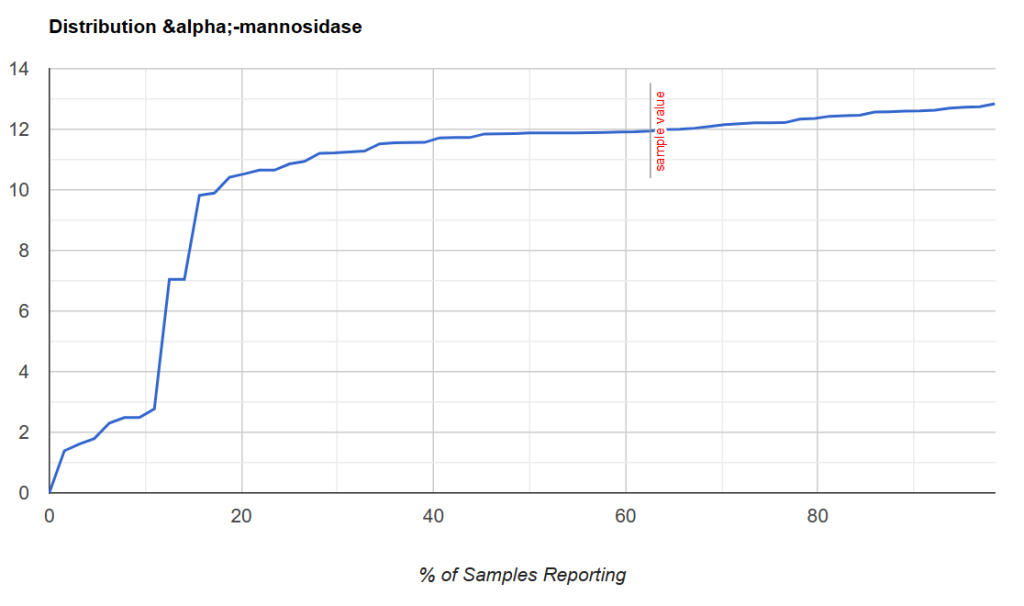
Thus it is possible to determine:
- If a person is likely deficient enough that supplementing enzymes via probiotic may be helpful
- We could infer dosages by the distance from the KM cutoff point.
Then we can proceed to apply this to a collection of retail probiotics products
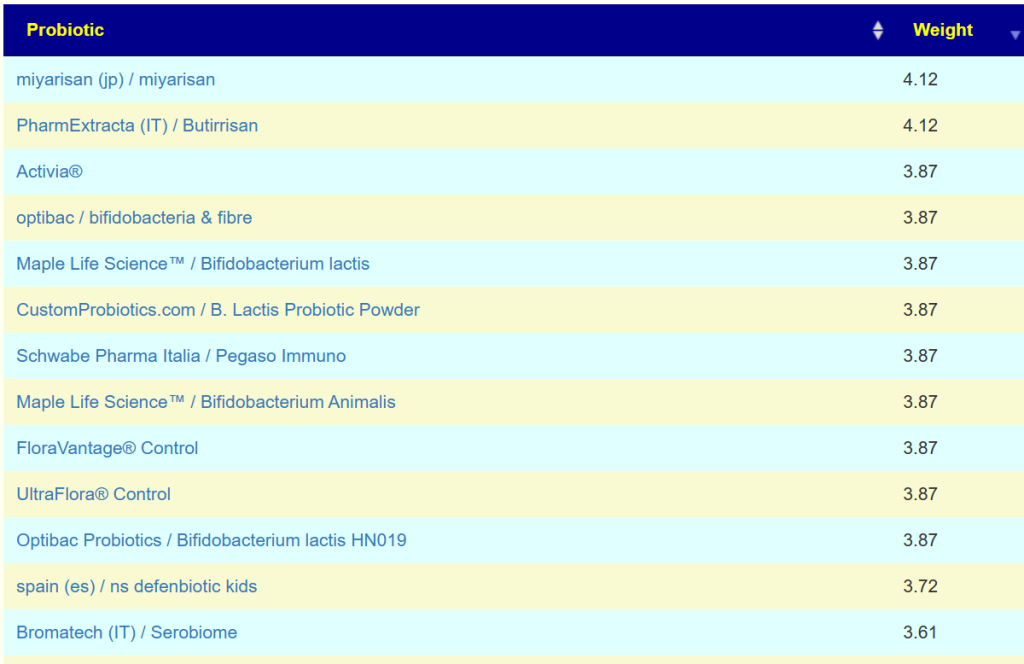
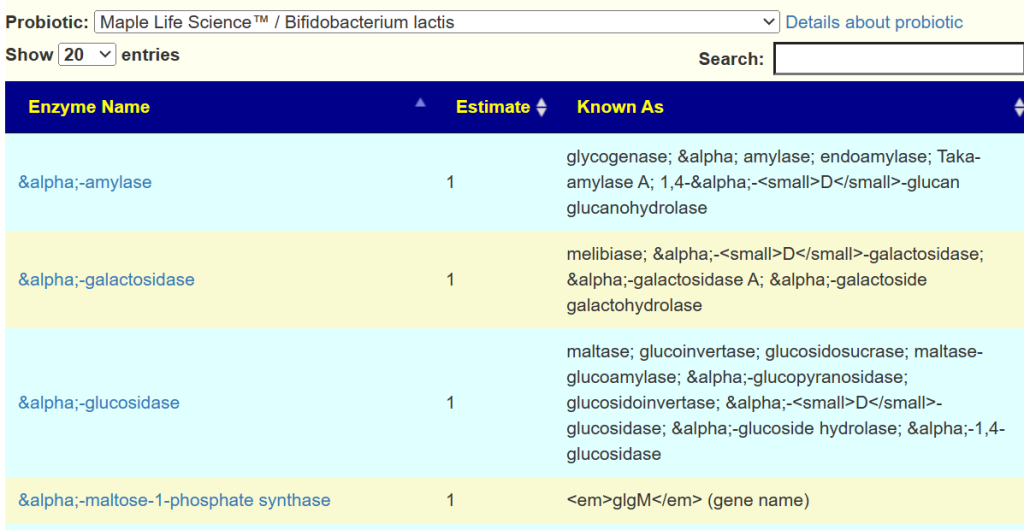
For example, Bifidobacterium Lactis was estimated to produce some 458 different enzymes.
Bottom Line
This approach does not try to “fix bacteria”, rather it tries to make sure that the fuel and oxygen need for the microbiome fire are there. Thus the bacteria issues resolve themselves! A very different way of trying to address microbiome dysfunction.
Recent Comments