Alternative Title: Climate Change and your guts!
This is a common symptom for both ME/CFS and Long COVID. This is reported often in samples uploaded. We examine if the data reaches our threshold for inclusion as defined in A new specialized selection of suggestions links. It does.
My default view is that this symptom is likely due to vesicular constriction/inflammation or “sticky blood” (which contains many possible coagulation issues). This results in slow blood flow causing issues with transferring heat to and from the body.
- temperature intolerance is generally placed under dysautonomia
Study Populations:
| Symptom | Reference | Study |
| Intolerance of extremes of heat and cold | 1159 | 54 |
- Bacteria Detected with z-score > 2.6: found 247 items, highest value was 8.8
- Enzymes Detected with z-score > 2.6: found 501 items, highest value was 6.7
- Compound Detected with z-score > 2.6: found No items
The number of bacteria and enzymes that are significant hints that it is a complex scenario, possibly with different subsets.
Interesting Significant Bacteria
All bacteria found significant had too low levels. The dominant order is Anaeroplasmatales and three significant genus are Holdemanella, Eubacterium, and Escherichia. Every one of the 247 bacteria found significant was LOW.
| Bacteria | Reference Mean | Study | Z-Score |
| Holdemanella biformis (species) | 3370 | 350 | 8.8 |
| Holdemanella (genus) | 3379 | 350 | 8.8 |
| Eubacterium (genus) | 2507 | 586 | 7.8 |
| Lysobacter deserti (species) | 29 | 11 | 6.5 |
| Anaerolineae (class) | 87 | 35 | 5.7 |
| Opitutae (class) | 163 | 46 | 5.5 |
| Puniceicoccaceae (family) | 158 | 44 | 5.5 |
| Legionellales (order) | 82 | 41 | 5.4 |
| Leptospiraceae (family) | 67 | 22 | 5.4 |
| Leptospira (genus) | 67 | 22 | 5.4 |
| Leptospirales (order) | 67 | 22 | 5.4 |
| Leptospira licerasiae (species) | 67 | 22 | 5.4 |
| Bifidobacterium cuniculi (species) | 79 | 26 | 5.3 |
| Lactiplantibacillus pentosus (species) | 116 | 22 | 5.3 |
| Puniceicoccales (order) | 110 | 37 | 5.3 |
| Legionellaceae (family) | 81 | 41 | 5.2 |
| Legionella (genus) | 81 | 41 | 5.2 |
| Anaeroplasmataceae (family) | 8102 | 20 | 5.2 |
| Anaeroplasma (genus) | 8102 | 20 | 5.2 |
| Anaeroplasmatales (order) | 8102 | 20 | 5.2 |
| Chloroflexi (phylum) | 128 | 55 | 5.1 |
| Cerasicoccus arenae (species) | 537 | 80 | 5.1 |
| Caldilineaceae (family) | 90 | 35 | 5.1 |
| Caldilinea (genus) | 90 | 35 | 5.1 |
| Caldilineales (order) | 90 | 35 | 5.1 |
| Caldilinea tarbellica (species) | 90 | 35 | 5.1 |
| Caldilineae (class) | 90 | 35 | 5.1 |
| Escherichia (genus) | 6016 | 1367 | 5.1 |
| Cerasicoccus (genus) | 312 | 60 | 5 |
Interesting Enzymes
Atypical distribution for enzymes in these studies, the number of highs and lows were of the same magnitude. 268 enzymes were low and 233 enzymes were high (see 2nd table) but none above our listing threshold of 5.0.
| Enzyme | Reference Mean | Study Mean | Z-Score |
| (S)-2-hydroxyglutarate:quinone oxidoreductase (1.1.5.13) | 2214 | 398 | 6.7 |
| (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase (succinate-forming) (4.1.3.30) | 1444 | 360 | 6.3 |
| propanoyl-CoA:oxaloacetate C-propanoyltransferase (thioester-hydrolysing, 1-carboxyethyl-forming) (2.3.3.5) | 1491 | 387 | 6.3 |
| propanoate:CoA ligase (AMP-forming) (6.2.1.17) | 1894 | 483 | 6.1 |
| D-galactaro-1,4-lactone lyase (ring-opening) (5.5.1.27) | 1698 | 393 | 5.8 |
| thiosulfate:ferricytochrome-c oxidoreductase (1.8.2.2) | 1381 | 272 | 5.4 |
| gamma-butyrobetainyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase (1.3.8.13) | 1400 | 441 | 5.4 |
| glutarate, 2-oxoglutarate:oxygen oxidoreductase ((S)-2-hydroxyglutarate-forming) (1.14.11.64) | 1299 | 297 | 5.4 |
| (E)-4-(trimethylammonio)but-2-enoyl-CoA:L-carnitine CoA-transferase (2.8.3.21) | 1386 | 440 | 5.4 |
| L-carnitinyl-CoA hydro-lyase [(E)-4-(trimethylammonio)but-2-enoyl-CoA-forming] (4.2.1.149) | 1444 | 321 | 5.4 |
| 2-dehydrotetronate isomerase (5.3.1.35) | 1406 | 482 | 5.3 |
| (2S,3S)-2-hydroxybutane-1,2,3-tricarboxylate hydro-lyase [(Z)-but-2-ene-1,2,3-tricarboxylate-forming] (4.2.1.79) | 1297 | 439 | 5.2 |
| 3-phenylpropanoate,NADH:oxygen oxidoreductase (2,3-hydroxylating) (1.14.12.19) | 581 | 142 | 5.2 |
| 1-aminocyclopropane-1-carboxylate aminohydrolase (isomerizing) (3.5.99.7) | 1431 | 416 | 5.2 |
| n/a (3.4.23.49) | 1307 | 326 | 5.2 |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate:NAD+ oxidoreductase (1.3.1.87) | 568 | 140 | 5.2 |
| ATP:2′-deoxyadenosine 5′-phosphotransferase (2.7.1.76) | 1974 | 344 | 5.2 |
| carotenoid beta-end group lyase (ring-opening) (5.5.1.19) | 1452 | 201 | 5.1 |
| RNA-3′-phosphate:RNA ligase (cyclizing, AMP-forming) (6.5.1.4) | 1220 | 316 | 5.1 |
| NTP:deoxycytidine 5′-phosphotransferase (2.7.1.74) | 1885 | 287 | 5.1 |
| L-threonate:NAD+ 2-oxidoreductase (1.1.1.411) | 1410 | 492 | 5.1 |
| succinyl-CoA:3-oxo-acid CoA-transferase (2.8.3.5) | 1307 | 363 | 5.1 |
| acyl-CoA,ferrocytochrome b5:oxygen oxidoreductase (6,7 cis-dehydrogenating) (1.14.19.3) | 1063 | 277 | 5 |
| L-carnitine:CoA ligase (AMP-forming) (6.2.1.48) | 1373 | 517 | 5 |
| hydrogen-sulfide:flavocytochrome c oxidoreductase (1.8.2.3) | 1165 | 161 | 5 |
| L-methionine methanethiol-lyase (deaminating; 2-oxobutanoate-forming) (4.4.1.11) | 1272 | 288 | 5 |
| L-carnitine,NAD(P)H:oxygen oxidoreductase (trimethylamine-forming) (1.14.13.239) | 1186 | 320 | 5 |
| 4-hydroxy-2-oxoglutarate glyoxylate-lyase (pyruvate-forming) (4.1.3.16) | 29 | 9 | 5 |

Bottom Line
Temperature intolerance is not an independently study topic. Existing studies are usually in the context of some other condition. This suggests that this is the first study on it’s microbiome.
- “Diabetes is often associated with orthostatic hypotension and temperature intolerance.” [2005]
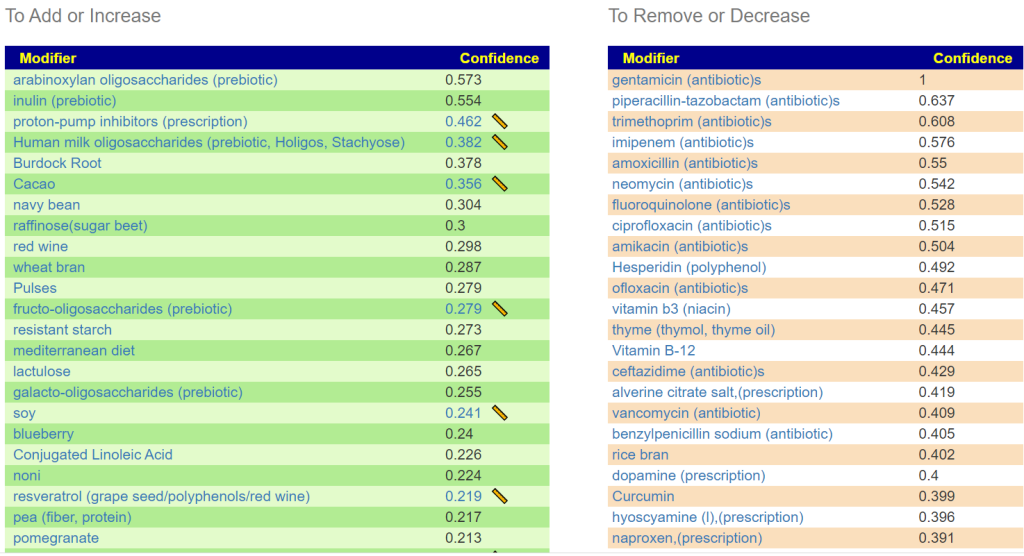
The suggestions below has some surprises on the to avoid list: Vitamins B-3, B-12, Curcumin, N-Acetyl Cysteine (NAC) sitting high in the list with antibiotics. The absence of probiotics in the to take suggestions is note worthy for those that view probiotics as ‘cure all’. Not listed, but conceptually worth considering, are the E.Coli probiotics (Symbioflor-2 or Mutaflor).
Recently I have see pea appear often and I recall that peas were served with most meals as a child. Peas has largely disappear from the western menu. I am starting to wonder if having pea soup 3 times a week would help a lot of people.
1 thought on “Special Study: Intolerance of Extremes of Heat and Cold”
Comments are closed.