This is a common symptom for many people. This is reported often in samples, and thus being examined if it reaches our threshold for inclusion as defined in A new specialized selection of suggestions links. It does. We are not being specific about the type of constipation.
Study Populations:
We have 2 symptom annotations that could be included
- Comorbid: Constipation and diarrhea (not explosions) – 29 only
- Comorbid: Constipation and Explosions (not diarrhea) – 11 only
- Immune Manifestations: Constipation – 9.7 max z-score (70)
- All of the above Constipation – 9.9 max z-score (83)
Taking all together we get 83 samples with an max z-score of 9.9
| Symptom | Reference | Study |
| Constipation | 1123 | 83 |
- Bacteria Detected with z-score > 2.6: found 123 items, highest value was 9.9
- Enzymes Detected with z-score > 2.6: found 511 items, highest value was 5.2
- Compound Detected with z-score > 2.6: found No items
Clearly some bacteria have strong associations, but the number of enzymes that are significant suggests that they may be more dimensions to the issue.
Previous studies have shown that the gut microbiota of constipated patients differs from healthy controls; however, many discrepancies exist in the findings, and no clear link has been confirmed between chronic constipation and changes in the gut microbiota.
Gut Microbiota Composition Changes in Constipated Women of Reproductive Age [2021]
The reason that I do not have Constipation on my US National Library of Medicine Studies list is that the studies are usually in the context of another microbiome altering condition. For example: Effects of Fermented Milk Containing Lacticaseibacillus paracasei Strain Shirota on Constipation in Patients with Depression [2021]
Interesting Significant Bacteria
Unusually bacteria was found significant with both high (13) and low (110). This far exceed the count expected as the false detection rate, so we should include and cite them.
| Bacteria | Reference Mean | Study | Z-Score |
| Prevotella copri (species) | 67087 | 7150 | 9.9 |
| Prevotella (genus) | 74786 | 17755 | 8 |
| Prevotellaceae (family) | 82267 | 29219 | 6.8 |
| Veillonella (genus) | 4003 | 2102 | 5 |
| Lactiplantibacillus pentosus (species) | 117 | 26 | 5 |
UNLIKE most of the other studies, we had a significant number of too many bacteria (far more than expected with a False Detection Rate).
| Bacteria | Reference Mean | Study | Z-Score |
| Tannerellaceae (family) | 24592 | 34890 | -3.6 |
| Parabacteroides (genus) | 24587 | 34665 | -3.6 |
| Porphyromonadaceae (family) | 26584 | 36605 | -3.4 |
| Bacteroides uniformis (species) | 25682 | 42170 | -3.2 |
| Bacteroidaceae (family) | 281711 | 337344 | -3.1 |
| Desulfosporosinus auripigmenti (species) | 18 | 28 | -3 |
| Hathewaya histolytica (species) | 2607 | 4093 | -2.8 |
| Bacteroides (genus) | 232904 | 276348 | -2.8 |
| Oscillospira (genus) | 22836 | 28653 | -2.8 |
| Anaerotruncus colihominis (species) | 1739 | 2436 | -2.8 |
| Anaerotruncus (genus) | 1834 | 2520 | -2.7 |
| Hathewaya (genus) | 2618 | 4058 | -2.7 |
| Parabacteroides merdae (species) | 7249 | 11459 | -2.6 |
There is some agreement with studies on these findings, but as cited above — results are not consistent in studies.
- Fecal bacterial microbiota in constipated patients before and after eight weeks of daily Bifidobacterium infantis 35624 administration [2022] , “Oscillospira, was the taxon most strongly correlated”
- “high abundances of Prevotella,… may be the main factors in curing constipation.[2020]
- ” studies have found that Bacteroides were more abundant in colonic mucosa of patients with chronic constipation” [2017]
Interesting Enzymes
As with bacteria we had both too few and too many.
| Enzyme | Reference Mean | Study Mean | Z-Score |
| UDP-N-acetyl-alpha-D-mannosamine:NAD+ 6-oxidoreductase (1.1.1.336) | 121077 | 167870 | -4.1 |
| GDP-alpha-D-mannose:2-O-alpha-D-mannosyl-1-phosphatidyl-1D-myo-inositol 6-alpha-D-mannosyltransferase (configuration-retaining) (2.4.1.346) | 43257 | 68972 | -4.1 |
| UDP-2-acetamido-3-amino-2,3-dideoxy-alpha-D-glucuronate:2-oxoglutarate aminotransferase (2.6.1.98) | 44942 | 68064 | -4.1 |
| acetyl-CoA:UDP-2-acetamido-3-amino-2,3-dideoxy-alpha-D-glucuronate N-acetyltransferase (2.3.1.201) | 67863 | 98332 | -4 |
| chondroitin AC lyase (4.2.2.5) | 62785 | 90081 | -3.8 |
| alpha-N-acetyl-D-glucosaminide N-acetylglucosaminohydrolase (3.2.1.50) | 170810 | 221135 | -3.7 |
| glycine:H-protein-lipoyllysine oxidoreductase (decarboxylating, acceptor-amino-methylating) (1.4.4.2) | 188544 | 239012 | -3.7 |
| L-leucyl-tRNALeu:[protein] N-terminal L-lysine/L-arginine leucyltransferase (2.3.2.6) | 129313 | 167839 | -3.7 |
| 5-phospho-alpha-D-ribose 1,2-cyclic phosphate 2-phosphohydrolase (alpha-D-ribose 1,5-bisphosphate-forming) (3.1.4.55) | 185457 | 234894 | -3.7 |
| (2E,6E)-farnesyl-diphosphate:isopentenyl-diphosphate farnesyltranstransferase (adding 5 isopentenyl units) (2.5.1.90) | 195115 | 245998 | -3.6 |
| Enzyme | Reference Mean | Study Mean | Z-Score |
| propanoyl-CoA:oxaloacetate C-propanoyltransferase (thioester-hydrolysing, 1-carboxyethyl-forming) (2.3.3.5) | 1509 | 500 | 5.2 |
| (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase (succinate-forming) (4.1.3.30) | 1458 | 500 | 4.9 |
| N-succinyl-LL-2,6-diaminoheptanedioate amidohydrolase (3.5.1.18) | 101418 | 72762 | 4.7 |
| ADP-alpha-D-glucose:alpha-D-glucose-1-phosphate 4-alpha-D-glucosyltransferase (configuration-retaining) (2.4.1.342) | 77187 | 49189 | 4.6 |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate:NAD+ oxidoreductase (1.3.1.87) | 580 | 183 | 4.5 |
| 3-phenylpropanoate,NADH:oxygen oxidoreductase (2,3-hydroxylating) (1.14.12.19) | 593 | 189 | 4.4 |
| D-tagatose 1,6-bisphosphate D-glyceraldehyde-3-phosphate-lyase (glycerone-phosphate-forming) (4.1.2.40) | 89078 | 62686 | 4.4 |
| UDP-N-acetyl-alpha-D-glucosamine:lipopolysaccharide N-acetyl-D-glucosaminyltransferase (2.4.1.56) | 1077 | 376 | 4.3 |
| D-aspartate:[beta-GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)]n ligase (ADP-forming) (6.3.1.12) | 87698 | 61735 | 4.3 |
| penicillin amidohydrolase (3.5.1.11) | 83364 | 57721 | 4.3 |
Bottom Line
While constipation is often assumed to be due too many of some bacteria, the evidence suggestions that not enough is the more likely cause. There appears to be no simple model or answer.
Prevotella copri will hopefully be available as a probiotic in a few year. There are two natural sources for P.Copri : Beer and Sauerkraut [2020], which may be an experiment for those that are prone to constipation..

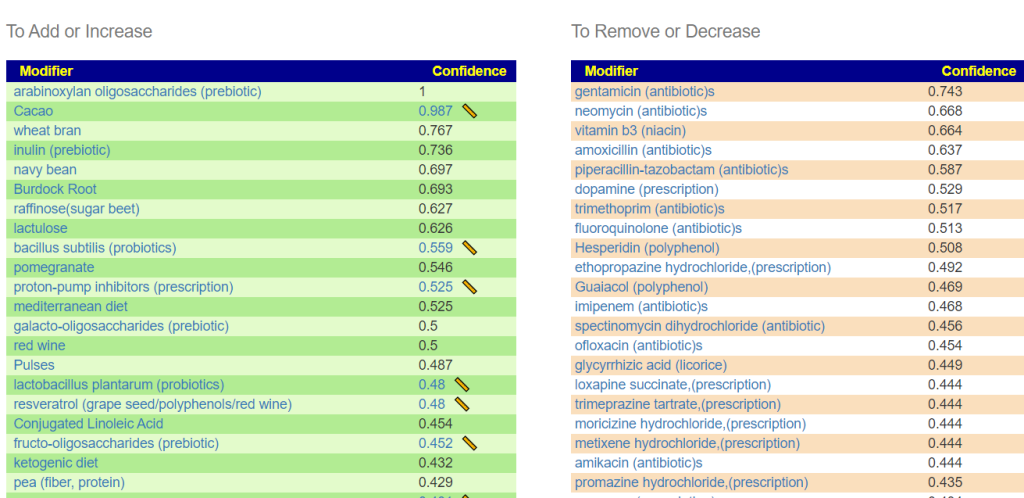
Looking at the suggestions — Constipation caused by Antibiotics!
This is not a “new discovery” — rather it appears to confirm that the mathematic model being used is reasonable and thus Dr. Artificial Intelligence suggestions are reasonable!
2 thoughts on “Special Studies: Constipation”
Comments are closed.