On April 14th, this new/revised feature was released. It determines shifts in these items by looking at the top and bottom 15% of people with these symptoms compared to the annotated sample population. This makes it easy to understand how significance is determined.
The basis is simple:
- We count the number of people with symptoms that have a percentile ranking below 15%ile or above 85%ile.
- If there is no association, then the numbers should be closed, i.e. 21 for each with the example below.
- If the numbers are different, we compute Chi2 , a statistical measure on the odds of it happening at random.
- A value of 6.635 means 1 in 100 (or P <.01)
- A value of 10.8 means 1 in 1000 (or P < 0.001)
- A value of 20 means 1 in 125,000 (or P < 0.000008)
- A value of 40 means 1 in 4,000,000,000 aka 4 billion (or P < 2.539629e-10)
- A Chi-Square Calculator for those interested.
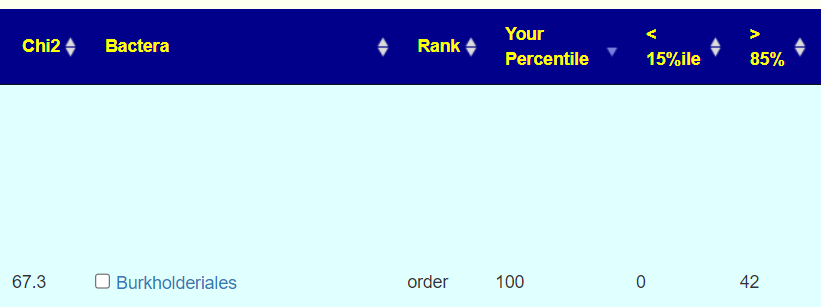
Rather than get into statistics, we show the common sense counts.
How to get there?
Upon logging with samples you will see this new menu item.
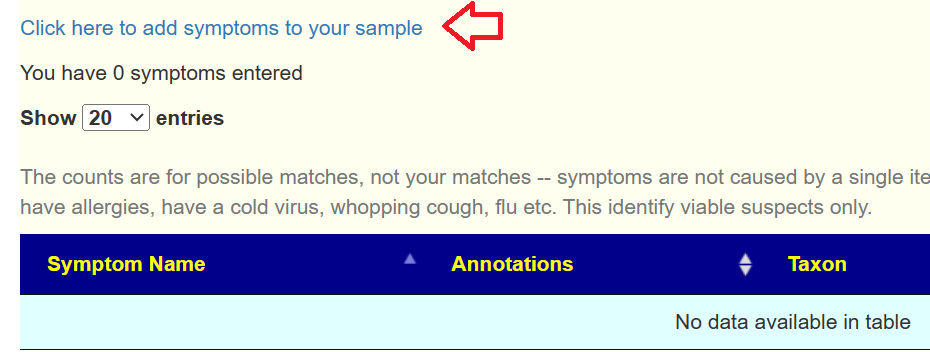
The next screen will matched against annotated symptoms for this sample. If you have no symptoms, this will be shown. You should add your symptoms via the link on this
If you have symptoms entered, then you will be shown a summary of what has been associated (according to samples from the lab you used)
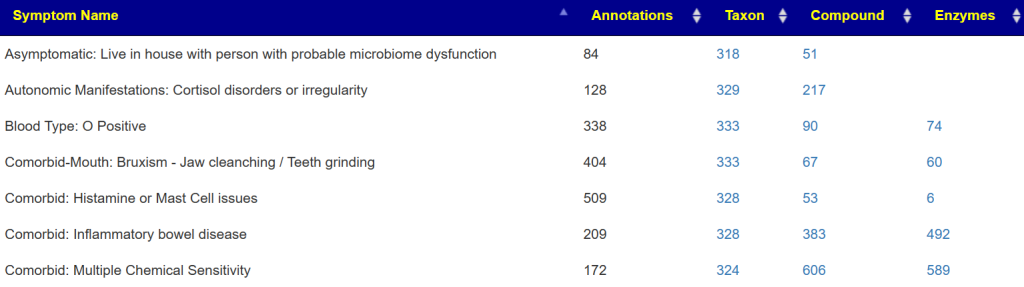
The right three columns are hyperlinked. The number of actual matches will be shown when you click the hyperlink. In some cases, many matches in other cases none.
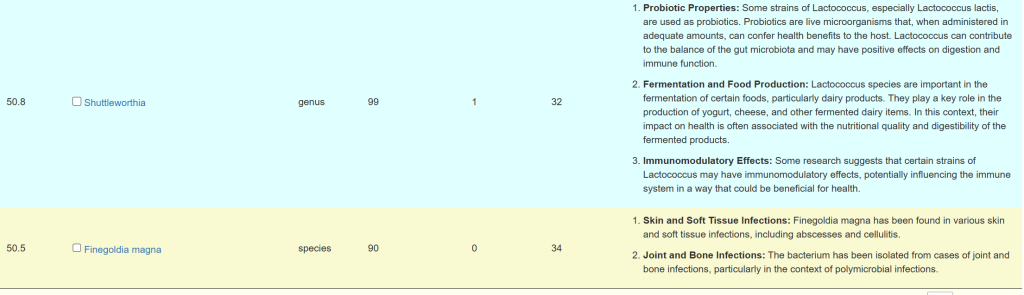
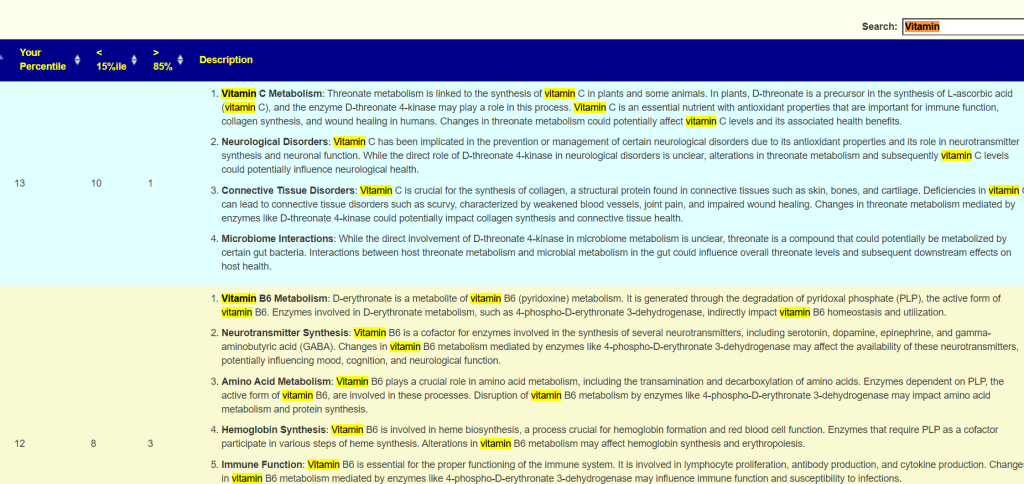
A general description is on the right. For Enzymes, typing “vitamin” in the search box. If the percentile is low, then you should consider supplementing with the vitamins listed. Why? you appears to be deficient in one or more enzymes that produces or uses it (as always, seek an opinion from an expert first)
Video Walk Thru
P.S. all of the bugs identified has been fixed.
Data Availability?
See https://citizenscience.microbiomeprescription.com/ for data. Kegg data on compounds and enzymes needs to be obtained from http://kegg.jp/ (licensing issue).
Recent Comments