This is a common symptom for many people. This is reported often in samples, and thus being examined if it reaches our threshold for inclusion as defined in A new specialized selection of suggestions links. It does. We are not being specific about the type of fatigue. Each person use their own subject definition of fatigue, thus we do not expect strong statistical associations (and do not get it!)
Study Populations:
I include values for Special Studies: General ME/CFS below
| Symptom | Reference | Study |
| General Fatigue | 1095 | 134 |
- Bacteria Detected with z-score > 2.6: found 158 items, highest value was 5.3 (ME/CFS was 6.6)
- Enzymes Detected with z-score > 2.6: found 410 items, highest value was 6.0 (ME/CFS was 4.5)
- Compound Detected with z-score > 2.6: found 67 items, highest values was -4/4 (ME/CFS was 3.1)
So we have a weaker bacteria signature but stronger enzymes and compound signature than ME/CFS. Many people marking one will mark the other… so get your sodium chloride crystals out!
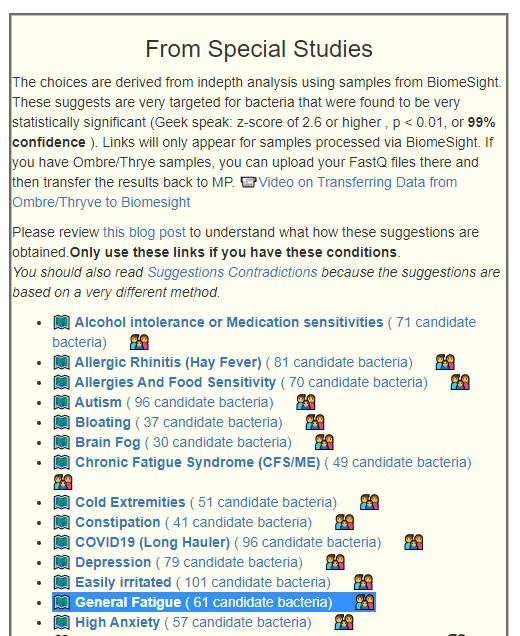
Interesting Significant Bacteria
All bacteria found significant had too low levels. The list of those with a z-score over 5 is small. Low Prevotella copri and Escherichia coli which appears on special studies on many co-morbid symptoms. The good news, is that there is work ongoing to produce a prevotella copri probiotic and several Escherichia coli probiotics are available.
We do see a few overgrowth These are seen only in some subsets.
| Bacteria | Reference Mean | Study | Z-Score |
| Lactiplantibacillus pentosus (species) | 114 | 22 | 5.3 |
| Prevotella copri (species) | 68098 | 21864 | 5.2 |
| Gammaproteobacteria (class) | 14382 | 5944 | 5.2 |
| Veillonella (genus) | 4022 | 2324 | 5.1 |
| Escherichia coli (species) | 829 | 196 | 5 |
Interesting Enzymes
Most enzymes found significant had too low levels. A few were higher (12 of 410), which are listed in a second table below
| Enzyme | Reference Mean | Study Mean | Z-Score |
| [cysteine desulfurase]-S-sulfanyl-L-cysteine:[molybdopterin-synthase sulfur-carrier protein]-Gly-Gly sulfurtransferase (2.8.1.11) | 5407 | 2420 | 6 |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate:NAD+ oxidoreductase (1.3.1.87) | 597 | 144 | 5.5 |
| 3-phenylpropanoate,NADH:oxygen oxidoreductase (2,3-hydroxylating) (1.14.12.19) | 611 | 148 | 5.5 |
| deoxyribocyclobutadipyrimidine pyrimidine-lyase (4.1.99.3) | 4311 | 1668 | 5.5 |
| tRNA-uridine65 uracil mutase (5.4.99.26) | 4201 | 1733 | 5.4 |
| S-adenosyl-L-methionine:23S rRNA (guanine2069-N7)-methyltransferase (2.1.1.264) | 5850 | 3067 | 5.3 |
| propanoyl-CoA:oxaloacetate C-propanoyltransferase (thioester-hydrolysing, 1-carboxyethyl-forming) (2.3.3.5) | 1575 | 501 | 5.2 |
| N4-acetylcytidine amidohydrolase (3.5.1.135) | 3083 | 1279 | 5.2 |
| ATP:D-tagatose 6-phosphotransferase (2.7.1.101) | 415 | 112 | 5.1 |
| (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase (succinate-forming) (4.1.3.30) | 1524 | 484 | 5.1 |
| acyl-CoA:sn-glycerol-3-phosphate 1-O-acyltransferase (2.3.1.15) | 3385 | 1438 | 5.1 |
| S-adenosyl-L-methionine:tRNA (cytidine32/uridine32-2′-O)-methyltransferase (2.1.1.200) | 3437 | 1390 | 5.1 |
| 2-(glutathione-S-yl)-hydroquinone:glutathione oxidoreductase (1.8.5.7) | 3263 | 1099 | 5.1 |
| galactitol-1-phosphate:NAD+ oxidoreductase (1.1.1.251) | 1042 | 292 | 5 |
| acetyl-CoA:[elongator tRNAMet]-cytidine34 N4-acetyltransferase (ATP-hydrolysing) (2.3.1.193) | 3430 | 1387 | 5 |
| S-adenosyl-L-methionine:23S rRNA (uracil747-C5)-methyltransferase (2.1.1.189) | 3379 | 1385 | 5 |
| ATP phosphohydrolase (ABC-type, thiamine-importing) (7.6.2.15) | 3637 | 1454 | 5 |
| [50S ribosomal protein L16]-L-Arg81,2-oxoglutarate:oxygen oxidoreductase (3R-hydroxylating) (1.14.11.47) | 3448 | 1440 | 5 |
| 7,8-dihydroneopterin 3′-triphosphate diphosphohydrolase (3.6.1.67) | 3719 | 1634 | 5 |
| ATP:N-acetyl-D-glucosamine 6-phosphotransferase (2.7.1.59) | 3383 | 1369 | 5 |
| Enzyme | Reference Mean | Study Mean | Z-Score |
| CTP:N,N’-diacetyllegionaminate cytidylyltransferase (2.7.7.82) | 59568 | 76504 | -2.9 |
| 1-phosphatidyl-1D-myo-inositol:a very-long-chain (2’R)-2′-hydroxy-phytoceramide phosphoinositoltransferase (2.7.1.227) | 39144 | 50749 | -2.9 |
| 1,5-anhydro-D-mannitol:NADP+ oxidoreductase (1.1.1.292) | 65010 | 81029 | -2.9 |
| alginate oligosaccharide 4-deoxy-alpha-L-erythro-hex-4-enopyranuronate-(1->4)-hexananopyranuronate lyase (4.2.2.26) | 79725 | 99196 | -2.8 |
| phosphatidylglycerophosphate phosphohydrolase (3.1.3.27) | 87820 | 105463 | -2.7 |
| chondroitin-sulfate-ABC endolyase (4.2.2.20) | 71773 | 88107 | -2.7 |
| chondroitin-sulfate-ABC exolyase (4.2.2.21) | 71773 | 88107 | -2.7 |
| arabinogalactan 4-beta-D-galactanohydrolase (3.2.1.89) | 107701 | 128121 | -2.7 |
| cephalosporin-C acetylhydrolase (3.1.1.41) | 86153 | 104494 | -2.7 |
| UDP-N-acetyl-alpha-D-glucosamine 4-epimerase (5.1.3.7) | 83349 | 100761 | -2.6 |
| N-sulfo-D-glucosamine sulfohydrolase (3.10.1.1) | 59634 | 73685 | -2.6 |
| 6-alpha-D-glucan 6-glucanohydrolase (3.2.1.11) | 64035 | 78529 | -2.6 |
| GDP-alpha-D-mannose:2-O-alpha-D-mannosyl-1-phosphatidyl-1D-myo-inositol 6-alpha-D-mannosyltransferase (configuration-retaining) (2.4.1.346) | 43744 | 55284 | -2.6 |
Interesting Compound
Unlike most of the special studies we have many compounds that are significant. I have listed the high and low in separate tables below. Spot checking most of these found no useful information. For Maltodextrin which becomes glucose would fit with fatigue — i.e. low sugar being produced.
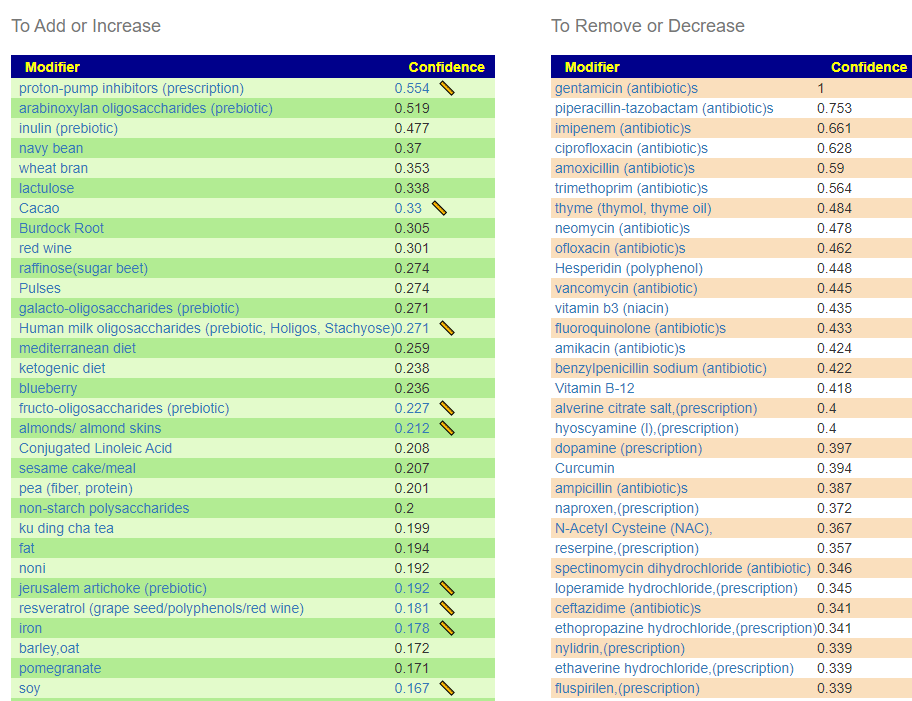
Of the low items, the following appear to be available as supplements and potentially could help with fatigue
- Maltodextrin
- Inosine
- Thymine (Vitamin B1)
- Cytosine (as N-acetylcysteine)
| Names | Z-Score |
| beta-D-Ribofuranose (C16639) | 4 |
| 1,2-Diacyl-3-alpha-D-glucosyl-sn-glycerol (C06364) | 3.8 |
| Coproporphyrin III (C05770) | 3.3 |
| Cys-Gly (C01419) | 3.2 |
| Deoxyinosine (C05512) | 3.2 |
| UDP-N-acetyl-alpha-D-glucosamine 3′-phosphate (C20245) | 3.2 |
| Inosine (C00294) | 3.1 |
| Carboxylate (C00060) | 3.1 |
| Thymine (C00178) | 2.9 |
| 3-(4-Hydroxyphenyl)pyruvate (C01179) | 2.9 |
| Acetoacetyl-CoA (C00332) | 2.8 |
| beta-D-Galactosyl-(1->3)-N-acetyl-D-galactosamine (C07278) | 2.8 |
| Prokaryotic ubiquitin-like protein (C21177) | 2.8 |
| dTDP (C00363) | 2.7 |
| tRNA with a 3′ CCA end (C19085) | 2.7 |
| Cytosine (C00380) | 2.7 |
| alpha-D-Aldose 1-phosphate (C00991) | 2.7 |
| 5′-O-Phosphonoadenylyl-(3′->5′)-adenosine (C22092) | 2.7 |
| Hexadecanoic acid (C00249) | 2.6 |
| Maltodextrin (C01935) | 2.6 |
| Hydroquinone (C15603) | 2.6 |
| Names | Z-Score |
| beta-D-Ribopyranose (C08353) | -4 |
| Chitobiose (C01674) | -3.6 |
| Deoxyadenosine (C00559) | -3.5 |
| Glutaredoxin (C07292) | -3 |
| tRNA(Leu) (C01645) | -3 |
| Lacto-N-biose (C06372) | -3 |
| alpha-D-Glucosamine 1-phosphate (C06156) | -3 |
| Pantetheine 4′-phosphate (C01134) | -2.9 |
| UDP-N-acetylmuramate (C01050) | -2.9 |
| L-Formylkynurenine (C02700) | -2.9 |
| tRNA uridine (C00868) | -2.9 |
| (S)-3-Hydroxybutanoyl-CoA (C01144) | -2.9 |
| Amino acid (C00045) | -2.8 |
| 1,2-Diacyl-sn-glycerol (C00641) | -2.8 |
| L-Glutamate 5-semialdehyde (C01165) | -2.8 |
| Taurine (C00245) | -2.8 |
| tRNA with a 3′ cytidine (C19078) | -2.8 |
| tRNA precursor (C02211) | -2.8 |
| Sucrose (C00089) | -2.8 |
| UTP (C00075) | -2.8 |
| beta-D-Glucose 1-phosphate (C00663) | -2.8 |
| UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine (C05892) | -2.7 |
| tRNA(Lys) (C01646) | -2.7 |
| L-Threonine (C00188) | -2.7 |
| 2-Succinylbenzoate (C02730) | -2.7 |
| Oxygen (C00007) | -2.7 |
| UDP-sugar (C05227) | -2.7 |
| Phenyl acetate (C15583) | -2.7 |
| Isopentenyl diphosphate (C00129) | -2.7 |
| Cyclomaltodextrin (C00973) | -2.7 |
Similarly, items that are too high are likely to be avoided, including the following
- Sucrose
- Taurine
- Threonine
All of these suggestions are theoretical based in the model. Some literature to consider that appears to confirm the above (a.k.a. cross-validation)
- A caffeine-maltodextrin mouth rinse counters mental fatigue [2018]
- Effect of inosine supplementation on aerobic and anaerobic cycling performance [1996]
- Thiamine and fatigue in inflammatory bowel diseases: an open-label pilot study [2013]
Bottom Line
II was not expecting much from this special study. I was pleased to see some suggestions being generated that can be implemented (or soon will be)
- Probiotics:
- Escherichia coli
- Prevotella copri
- Supplements
- Maltodextrin
- Inosine
- Thymine (Vitamin B1)
- Cytosine (as N-acetylcysteine)
In looking at the suggestions below, remember we are using two very different models. Above we use KEGG data to identify what the bacteria are producing (the items going to the farmer’s market). Below, we use what has been reported to influence the population of the bacteria that we are too low in (i.e. “Fertilizer”)
1 thought on “Special Studies: General Fatigue”
Comments are closed.