Author: Research
Symptoms with Ability to Predict from Microbiome Results
The table below is based on Biomesight samples which has been annotated with self-reporting symptoms. A factor of 16 means no ability to predict.
- A value of 20 indicates about a 56% chance of correctly predicting both With and Without
- A value of 25 indicates about a 66% chance of correctly predicting both With and Without
- A value of 30 indicates about a 70% chance of correctly predicting both With and Without
The chance can increase significantly (90+%) when you are willing to have a higher percentage of false positive.
The Table
| Symptom Name | Factor |
| Neurological-Sleep: Sleep Apnea | 30.42 |
| Comorbid: Sugars cause sleep or cognitive issues | 29.39 |
| Comorbid: Panic Attacks | 29.30 |
| Comorbid: Carbohydrate intolerance | 28.89 |
| Official Diagnosis: Hypercholesterolemia (High Cholesterol) | 28.51 |
| DePaul University Fatigue Questionnaire : Fever & Chills | 28.14 |
| Neurological: Disorientation | 27.99 |
| Comorbid: Sleep Apnea Diagnosis | 27.83 |
| Comorbid: Salicylate sensitive | 27.49 |
| Pain: Eye pain | 27.46 |
| Neurological: Seasonal Affective Disorder (SAD) | 27.39 |
| Comorbid-Mouth: Gingivits / Gum Disease | 27.31 |
| Condition: Generalized anxiety disorder | 27.21 |
| Condition: Post-Traumatic Stress Disorder | 27.09 |
| DePaul University Fatigue Questionnaire : Night sweats | 27.05 |
| Neurological-Sleep: Night Sweats | 26.96 |
| Autonomic Manifestations: exertional dyspnea | 26.96 |
| Comorbid: Constipation and Explosions (not diarrohea) | 26.46 |
| Autonomic Manifestations: cardiac arrhythmias | 26.39 |
| Neurological-Sleep: Vivid Dreams/Nightmares | 26.39 |
| Autism: High Functioning | 26.32 |
| DePaul University Fatigue Questionnaire : Tense muscles | 26.27 |
| Infection: Lyme | 26.26 |
| Comorbid: Mold Sensitivity / Exposure | 26.26 |
| Autonomic Manifestations: Cortisol disorders or irregularity | 26.22 |
| Onset: less than 32 years since onset | 26.14 |
| DePaul University Fatigue Questionnaire : Frequently get words or numbers in the wrong order | 26.05 |
| Pain: Sensitivity to pain | 26.02 |
| Blood Type: B Positive | 25.99 |
| Other: Sensitivity to vibrations | 25.97 |
| Autonomic Manifestations: bladder dysfunction | 25.96 |
| Joint: Stiffness and swelling | 25.95 |
| Comorbid: Raynaud’s syndrome (Skin discoloration) | 25.93 |
| DePaul University Fatigue Questionnaire : Confusion/disorientation | 25.87 |
| DePaul University Fatigue Questionnaire : New trouble with math | 25.87 |
| Immune Manifestations: Thick blood / Hypercoagulation | 25.82 |
| DePaul University Fatigue Questionnaire : Concern with driving | 25.82 |
| DePaul University Fatigue Questionnaire : Weight change | 25.79 |
| Pain: Myofascial pain | 25.77 |
| Comorbid-Mouth: Mouth Sores | 25.76 |
| DePaul University Fatigue Questionnaire : Chilled or shivery | 25.74 |
| Official Diagnosis: Fibromyalgia | 25.73 |
| Neuroendocrine: Feeling like you have a high temperature | 25.65 |
| Neurological-Sleep: Chaotic diurnal sleep rhythms (Erratic Sleep) | 25.54 |
| Neuroendocrine Manifestations: Excessive adrenaline | 25.52 |
| Neurological: fasciculations | 25.50 |
| Age: 60-70 | 25.48 |
| Autonomic: Inability to tolerate an upright position | 25.39 |
| Comorbid: Mood Swings | 25.38 |
| Comorbid: Inflammatory bowel disease | 25.37 |
| Autonomic Manifestations: urinary frequency dysfunction | 25.32 |
| Autonomic: Dizziness or fainting | 25.31 |
| DePaul University Fatigue Questionnaire : Depression | 25.30 |
| Neuroendocrine: Temperature fluctuations throughout the day | 25.29 |
| Comorbid: Restless Leg | 25.29 |
| Comorbid: Snoring (NOT Sleep Apnea | 25.29 |
| DePaul University Fatigue Questionnaire : Nausea | 25.28 |
| Official Diagnosis: Attention deficit hyperactivity disorder (ADHD) | 25.24 |
| DePaul University Fatigue Questionnaire : Feeling like you have a temperature | 25.22 |
| Infection: Human Herpesvirus 6 (HHV6) | 25.22 |
| Neurological: High degree of Empathy before onset | 25.21 |
| DePaul University Fatigue Questionnaire : Chemical sensitivity | 25.19 |
| DePaul University Fatigue Questionnaire : Sore Throat | 25.17 |
| Neuroendocrine Manifestations: abnormal appetite | 25.17 |
| General: Anhedonia (inability to feel pleasure) | 25.13 |
| Neurological-Vision: Blurred Vision | 25.09 |
| Neurological-Vision: photophobia (Light Sensitivity) | 25.02 |
| Immune Manifestations: tender lymph nodes | 24.97 |
| DePaul University Fatigue Questionnaire : Upset stomach | 24.95 |
| Pain: Aching of the eyes or behind the eyes | 24.86 |
| Neurological: Confusion | 24.86 |
| Neuroendocrine Manifestations: marked weight change | 24.83 |
| DePaul University Fatigue Questionnaire : Dizziness | 24.74 |
| Neuroendocrine Manifestations: subnormal body temperature | 24.68 |
| DePaul University Fatigue Questionnaire : Racing heart | 24.64 |
| Immune: Tender / sore lymph nodes | 24.62 |
| Comorbid: Fibromyalgia | 24.58 |
| Comorbid: Migraine | 24.54 |
| General: Myalgia (pain) | 24.54 |
| Neurocognitive: Feeling disoriented | 24.51 |
| Official Diagnosis: Depression | 24.34 |
| Neurocognitive: Unable to focus vision and/or attention | 24.34 |
| Neurological: Neuropathy | 24.34 |
| DePaul University Fatigue Questionnaire : Abnormal sensitivity to light | 24.32 |
| Physical: Good Air Quality | 24.27 |
| Official Diagnosis: Gastroesophageal reflux disease (GERD) | 24.27 |
| Joint: Tenderness | 24.26 |
| Physical: Tonsils removed | 24.24 |
| Comorbid: Methylation issues (MTHFR) | 24.23 |
| DePaul University Fatigue Questionnaire : Tingling feeling | 24.20 |
| Other: Sensitivity to mold | 24.17 |
| Neurological: emotional overload | 24.17 |
| Official Diagnosis: COVID19 (Fully Recovered) | 24.15 |
| Comorbid-Mouth: Dry Mouth | 24.13 |
| Physical: Steps Per Day 8000-16000 | 24.12 |
| Autonomic: Shortness of breath | 24.04 |
| Neuroendocrine: Lack of appetite | 24.03 |
| DePaul University Fatigue Questionnaire : Slow to react | 23.99 |
| Neuroendocrine Manifestations: loss of adaptability | 23.93 |
| DePaul University Fatigue Questionnaire : Rash | 23.90 |
| Autonomic: Irregular heartbeats | 23.87 |
| Physical: Organic Diet | 23.80 |
| Immune: Viral infections with prolonged recovery periods | 23.79 |
| Autonomic Manifestations: light-headedness | 23.75 |
| DePaul University Fatigue Questionnaire : Trouble expressing thoughts | 23.70 |
| DePaul University Fatigue Questionnaire : Feel unsteady on feet | 23.61 |
| Official Diagnosis: Mast Cell Dysfunction | 23.61 |
| Onset: less than 02 years since onset | 23.60 |
| Neurological: Difficulty reading | 23.59 |
| Neurological: Difficulty processing information (Understanding) | 23.57 |
| Immune Manifestations: Inflammation of skin, eyes or joints | 23.55 |
| Neuroendocrine Manifestations: sweating episodes | 23.42 |
| Neurological: Executive Decision Making (Difficulty making) | 23.39 |
| DePaul University Fatigue Questionnaire : Absent-mindedness | 23.34 |
| General: Heavy feeling in arms and legs | 23.32 |
| Infection: Epstein-Barr virus | 23.30 |
| DePaul University Fatigue Questionnaire : Abdomen pain | 23.29 |
| Neuroendocrine Manifestations: Air Hunger | 23.25 |
| Pain: Joint pain | 23.25 |
| Neurological-Sleep: Inability for deep (delta) sleep | 23.24 |
| Autonomic Manifestations: Orthostatic intolerance | 23.20 |
| DePaul University Fatigue Questionnaire : Does physical activity make you feel better | 23.18 |
| Comorbid: Multiple Chemical Sensitivity | 23.15 |
| Post-exertional malaise: Next-day soreness after everyday activities | 23.13 |
| Official Diagnosis: Autoimmune Disease | 23.09 |
| DePaul University Fatigue Questionnaire : Frequently loose train of thought | 23.09 |
| Immune Manifestations: Mucus in the stool | 23.08 |
| Condition: Non-Celiac Gluten Sensitivity | 23.05 |
| DePaul University Fatigue Questionnaire : Difficulty retaining information | 23.04 |
| DePaul University Fatigue Questionnaire : Slowness of thought | 23.01 |
| Neurological: Dysautonomia | 22.95 |
| DePaul University Fatigue Questionnaire : Difficulty reasoning things out | 22.94 |
| Comorbid-Mouth: TMJ / Dysfunction of the temporomandibular joint syndrome | 22.93 |
| DePaul University Fatigue Questionnaire : Eye pain | 22.92 |
| DePaul University Fatigue Questionnaire : Pain in Multiple Joints without Swelling or Redness | 22.90 |
| Neuroendocrine Manifestations: Painful menstrual periods | 22.90 |
| Neurological: Myoclonic jerks or seizures | 22.89 |
| DePaul University Fatigue Questionnaire : Shortness of breath | 22.88 |
| Autonomic: Ocassional Tachycardia (Rapid heart beat) | 22.86 |
| Immune Manifestations: recurrent flu-like symptoms | 22.86 |
| Neurological: Cognitive/Sensory Overload | 22.84 |
| DePaul University Fatigue Questionnaire : Difficulty following things | 22.84 |
| Autonomic Manifestations: Postural orthostatic tachycardia syndrome (POTS) | 22.83 |
| Immune: Chronic Sinusitis | 22.83 |
| DePaul University Fatigue Questionnaire : Temperature lower than normal | 22.82 |
| DePaul University Fatigue Questionnaire : Impaired Memory & concentration | 22.76 |
| Comorbid: Hypothyroidism | 22.74 |
| DePaul University Fatigue Questionnaire : Hot or Cold spells | 22.72 |
| Official Diagnosis: Autism | 22.72 |
| DePaul University Fatigue Questionnaire : Muscle weakness | 22.68 |
| Neuroendocrine Manifestations: Muscle weakness | 22.64 |
| Autonomic Manifestations: nausea | 22.62 |
| Autonomic: Nausea | 22.59 |
| Age: 30-40 | 22.59 |
| Blood Type: O Positive | 22.58 |
| DePaul University Fatigue Questionnaire : Mood swings | 22.58 |
| Sleep: Need to nap daily | 22.57 |
| Neuroendocrine: Feeling hot or cold for no reason | 22.53 |
| DePaul University Fatigue Questionnaire : Sensitivity to Alcohol | 22.52 |
| DePaul University Fatigue Questionnaire : Blurred Vision | 22.49 |
| Onset: less than 04 years since onset | 22.47 |
| Physical: Amalgam fillings | 22.43 |
| Neuroendocrine Manifestations: Paraesthesia (tingling burning of skin) | 22.42 |
| Post-exertional malaise: Mentally tired after the slightest effort | 22.40 |
| Neurological-Vision: inability to focus eye/vision | 22.38 |
| Immune: Recurrent Sore throat | 22.38 |
| DePaul University Fatigue Questionnaire : Difficulty staying asleep | 22.37 |
| DePaul University Fatigue Questionnaire : Need to have to focus on one thing at a time | 22.37 |
| DePaul University Fatigue Questionnaire : Ringing in the Ears | 22.34 |
| Immune Manifestations: Diarrhea | 22.26 |
| General: Headaches | 22.25 |
| Immune Manifestations: Hair loss | 22.25 |
| DePaul University Fatigue Questionnaire : Difficulty comprehending Information | 22.24 |
| Neuroendocrine Manifestations: Rapid muscular fatiguability | 22.24 |
| Post-exertional malaise: Worsening of symptoms after mild mental activity | 22.24 |
| Physical: Breastfed | 22.23 |
| Physical: Long term (chronic) stress | 22.22 |
| DePaul University Fatigue Questionnaire : Headaches | 22.21 |
| Physical: Steps Per Day 2000-4000 | 22.21 |
| Neuroendocrine: Lost or gained weight without trying | 22.19 |
| Autonomic Manifestations: irritable bowel syndrome | 22.18 |
| Post-exertional malaise: Physically drained or sick after mild activity | 22.15 |
| General: Depression | 22.15 |
| Physical: Pets | 22.14 |
| Neurological: Joint hypermobility | 22.12 |
| DePaul University Fatigue Questionnaire : Forgetting what you are trying to say | 22.10 |
| Neurological-Audio: hypersensitivity to noise | 22.04 |
| Comorbid: High Anxiety | 22.02 |
| Immune Manifestations: Inflammation (General) | 21.97 |
| Age: 50-60 | 21.94 |
| Onset: 2000-2010 | 21.89 |
| Immune: Flu-like symptoms | 21.85 |
| DePaul University Fatigue Questionnaire : Anxiety/tension | 21.78 |
| Sleep: Waking up early in the morning (e.g. 3 AM) | 21.76 |
| Post-exertional malaise: Muscle fatigue after mild physical activity | 21.75 |
| Post-exertional malaise: Physically tired after minimum exercise | 21.70 |
| Comorbid: Constipation and Diarrohea (not explosions) | 21.70 |
| DePaul University Fatigue Questionnaire : Muscle Pain (i.e., sensations of pain or aching in your muscles. This does not include weakness or pain in other areas such as joints) | 21.69 |
| Comorbid-Mouth: Bruxism – Jaw cleanching / Teeth grinding | 21.65 |
| Condition: Acne | 21.63 |
| Immune: Sensitivity to smell/food/medication/chemicals | 21.61 |
| Blood Type: A Positive | 21.61 |
| Immune Manifestations: Abdominal Pain | 21.59 |
| Physical: Northern European | 21.58 |
| Pain: Pain or aching in muscles | 21.54 |
| Neuroendocrine Manifestations: intolerance of extremes of heat and cold | 21.52 |
| General: Sinus issues with headaches | 21.48 |
| Post-exertional malaise: Post-exertional malaise | 21.48 |
| Post-exertional malaise: Rapid muscular fatigability, | 21.44 |
| Neuroendocrine: Cold limbs (e.g. arms, legs hands) | 21.42 |
| Immune Manifestations: Chronic Flatus / Flatulence / gas | 21.39 |
| DePaul University Fatigue Questionnaire : Allergies | 21.39 |
| DePaul University Fatigue Questionnaire : Does physical activity make you feel worse | 21.37 |
| Onset: less than 16 years since onset | 21.35 |
| Neuroendocrine Manifestations: Dry Eye (Sicca or Sjogren Syndrome) | 21.35 |
| Neurocognitive: Slowness of thought | 21.35 |
| Neurological: Word-finding problems | 21.30 |
| Neurological: Impairment of concentration | 21.29 |
| Immune Manifestations: new food sensitivities | 21.28 |
| Post-exertional malaise: Worsening of symptoms after mild physical activity | 21.27 |
| Physical: Steps Per Day 4000-8000 | 21.26 |
| Immune Manifestations: Alcohol Intolerant | 21.25 |
| Physical: Steps Per Day < 2000 | 21.24 |
| Condition: ME/CFS without IBS | 21.23 |
| DePaul University Fatigue Questionnaire : Need to nap during each day | 21.23 |
| DePaul University Fatigue Questionnaire : Poor Appetite | 21.23 |
| DePaul University Fatigue Questionnaire : Fatigue | 21.21 |
| Physical: Eastern European | 21.21 |
| Neurocognitive: Can only focus on one thing at a time | 21.20 |
| Condition: ME/CFS with IBS | 21.15 |
| Neurocognitive: Problems remembering things | 21.13 |
| DePaul University Fatigue Questionnaire : Difficulty recalling information | 21.10 |
| DePaul University Fatigue Questionnaire : Difficulty falling asleep | 21.08 |
| Sleep: Daytime drowsiness | 21.07 |
| Neurocognitive: Absent-mindedness or forgetfulness | 21.07 |
| Post-exertional malaise: Rapid cognitive fatigability, | 21.05 |
| Onset: Sudden | 21.03 |
| DePaul University Fatigue Questionnaire : Easily irritated | 21.03 |
| DePaul University Fatigue Questionnaire : Walking up early in the morning (e.g. 3AM) | 21.03 |
| Immune Manifestations: medication sensitivities. | 21.01 |
| Neurological-Audio: Tinnitus (ringing in ear) | 20.98 |
| Comorbid: Small intestinal bacterial overgrowth (SIBO) | 20.96 |
| Physical: Work-Sitting | 20.96 |
| Neuroendocrine: Alcohol intolerance | 20.91 |
| Neurological-Sleep: Insomnia | 20.91 |
| Onset: Gradual | 20.90 |
| Neurocognitive: Difficulty understanding things | 20.84 |
| Autonomic Manifestations: palpitations | 20.80 |
| Neurological: Short-term memory issues | 20.71 |
| DePaul University Fatigue Questionnaire : Post-exertional malaise, feeling worse after doing activities that require either physical or mental exertion | 20.52 |
| Autonomic: Heart rate increase after standing | 20.51 |
| Immune Manifestations: Bloating | 20.51 |
| DePaul University Fatigue Questionnaire : Unrefreshing Sleep, that is waking up feeling tired | 20.49 |
| DePaul University Fatigue Questionnaire : Difficulty finding the right word | 20.46 |
| Comorbid: Histamine or Mast Cell issues | 20.41 |
| Gender: Female | 20.35 |
| Post-exertional malaise: Inappropriate loss of physical and mental stamina, | 20.33 |
| Neurocognitive: Difficulty expressing thoughts | 20.27 |
| Post-exertional malaise: Difficulty reading after mild physical or mental activity | 20.25 |
| Official Diagnosis: Chronic Fatigue Syndrome (CFS/ME) | 20.20 |
| Post-exertional malaise: General | 20.19 |
| Official Diagnosis: COVID19 (Long Hauler) | 20.18 |
| Official Diagnosis: Allergic Rhinitis (Hay Fever) | 20.06 |
| Neuroendocrine Manifestations: Poor gut motility | 20.04 |
| Sleep: Problems staying asleep | 20.03 |
| Onset: 2010-2020 | 20.01 |
| Neuroendocrine Manifestations: worsening of symptoms with stress. | 19.94 |
| Neurocognitive: Brain Fog | 19.88 |
| Immune Manifestations: general malaise | 19.84 |
| Sleep: Problems falling asleep | 19.75 |
| Neuroendocrine Manifestations: cold extremities | 19.68 |
| Official Diagnosis: Irritable Bowel Syndrome | 19.64 |
| Neurocognitive: Difficulty paying attention for a long period of time | 19.57 |
| Immune Manifestations: Constipation | 19.42 |
| Age: 20-30 | 19.37 |
| Age: 40-50 | 18.92 |
| Sleep: Unrefreshed sleep | 18.84 |
| General: Fatigue | 18.39 |
| Gender: Male | 18.29 |
This is part of research in progress and intended to indicate the degree that the microbiome may contribute to various symptoms. The above was based on testing models against this data to determine how often it predicted correctly.
Work is in progress to generate suggestions to moderate the microbiome away from these symptoms.
For a list of the genus for each, see Citizen Science Symptoms To Genus Special Studies. Using 84+%ile for high to be deemed a match, and 16-% for low to be deemed a match has produced good results,
Food/supplements recommendations for increasing serotonin(5-HT)
This was a request from a reader and I will do a direct approach Items reported from studies.
Items from Published Studies
- Tryptophan: This is an amino acid that the body converts into serotonin. It’s found in foods like turkey, cheese, and nuts, but it’s also available as a supplement.
- “Tryptophan, an indispensable amino acid, is needed to support protein deposition, and the synthesis of the neurotransmitter serotonin,”[2022]
- “Tryptophan is the only precursor of serotonin and mediates serotonergic activity in the brain. Previous studies have shown that the administration of tryptophan or tryptophan depletion significantly alters cognition, mood and anxiety. ” [2017]
- “Supplementation with B vitamins and magnesium has an influence on the Trp concentration.” [Tryptophan status in autism spectrum disorder and the influence of supplementation on its level, 2017]
- Tryptophan supplementation and serotonin function: genetic variations in behavioural effects [2018]
- 5-HTP (5-Hydroxytryptophan): This is a by-product of tryptophan and is directly converted into serotonin in the body. It’s often used as a supplement for mood enhancement.
- Tryptophan-enriched diet or 5-hydroxytryptophan supplementation given in a randomized controlled trial impacts social cognition on a neural and behavioral level [2021]
- 5-Hydroxytryptophan (5-HTP): Natural Occurrence, Analysis, Biosynthesis, Biotechnology, Physiology and Toxicology [2020]
- “An excess of 5-HTP may be responsible for serotonin syndrome (see Section 8.2.1) and an excessive treatment was found to be associated with severe side effects, including behavioral disturbances, abnormal mental functions, and intolerance.”
- St. John’s Wort: Often used for depression, St. John’s Wort is believed to affect serotonin levels, although its exact mechanism is not fully understood.
- Extractive from Hypericum ascyron L promotes serotonergic neuronal differentiation in vitro [2018]
- Mechanism of action of St John’s wort in depression : what is known? [2003]
- “St John’s wort extract leads to a downregulation of beta-adrenergic receptors and an upregulation of serotonin 5-HT(2) receptors in the rat frontal cortex”
SAMe (S-adenosylmethionine): This is a compound naturally found in the body that is thought to enhance neurotransmitter production, including serotonin.- “Use of SAMe elicited no significant adverse effects beyond placebo, however it was implicated in one case of serotonin syndrome-like symptoms.” [2018]
- Omega-3 Fatty Acids: Found in fish oil and certain plant oils, omega-3 fatty acids are linked to improved mental health and mood regulation, possibly through influencing serotonin pathways.
- Effect of the dietary intake of fish oil on psycho-social behavioral disorder caused by social-defeat stress [2022] “This improvement could be explained by the increase in serotonin synthesis in the hippocampus.”
- “Fish-oil supplementation decreases Indoleamine-2,3-Dioxygenase expression and increases hippocampal serotonin levels in the LPS depression model” [2020]
- Vitamin B6 and B12: These vitamins are important for serotonin production. A deficiency in these vitamins can lead to reduced serotonin levels.
- “vitamin B6, a cofactor in the tryptophan-serotonin pathway critical to mood regulation.” [2023]
- Link between higher levels of homocysteine and depression [2012]
- “Homocystcinc is reconstituted into methionine, which is essential to the production of neurotransmitters such as serotonin and dopamine which elevate mood. This reconstitution requires B-group vitamins, especially folic acid and B12. “
- Magnesium: This mineral plays a role in many biochemical reactions in the body and has been suggested to have a mood-stabilizing effect, possibly by influencing serotonin.
- Effect of subacute manganese feeding on serotonin metabolism in the rat [1978]
- Probiotics: Some research suggests that gut health can impact serotonin levels, as a significant amount of this neurotransmitter is produced in the gut.
- lactobacillus plantarum
- Lactobacillus rhamnosus GG
- Mutaflor (Escherichia coli strain Nissle 1917)
Foods
- Foods with Tryptophan: “chicken, soya beans, cereals, tuna, nuts and bananas’ may serve as an alternative to improve mood and cognition. ‘ [2013]
- Withania somnifera ( ashwagandha)
- “subjects showed significant increases in serum serotonin, gastrin,” [2024]
- kiwifruit
- ” treatments increased urinary concentration of the serotonin metabolite” [2023]
- poly-γ-glutamic acid with vitamin B6
- “a greater increase in the group C intervention (4.59 ± 38.5 ng/mL) in serum serotonin concentrations’ [2021]
Vitamin D has no effect (multiple studies)
Technical Note: Applying Studies to Individuals Part I
In this post I will show the results of a series of experiments using the results of prior posts, . The philosophical question being asked is this “How useful is a result showing that genus X mean is higher with a condition then without for screening individuals?” (i.e. with statistical significance of P < 0.01 or better).
Often studies will provide a statement such as the one shown below.
Men with higher VFA harbored a smaller relative abundance of Blautia and Bifidobacterium (P for trend: 0.003 and 0.021, respectively),
Blautia genus associated with visceral fat accumulation in adults 20-76 years of age [2019]
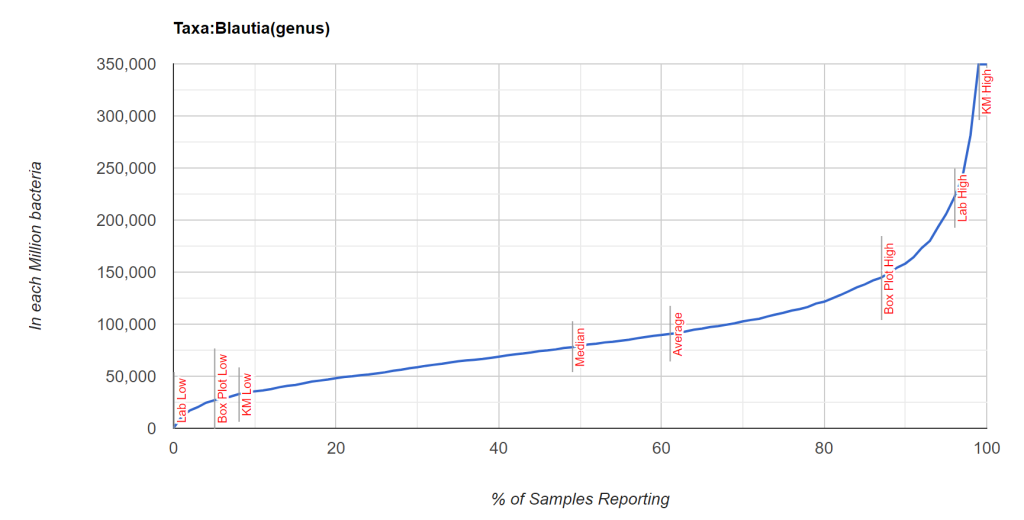
While it is possible to compute the t-score from p < 0.003 [assuming sample size of 100 was used, then 2.871] and then a priori apply it to the mean and standard deviation of population for a specific lab (per million) [mean: 89,844, standard deviation: 60,542] and get a proxy mean for male VFA = mean – 2.871 * StdDev/ 10 => 43,160.
We do not have an answer for the probability of a sample with 50,000 or 20,000 units. We could assume we have a normal distribution but that is an naïve assumption. A normal distribution would have average and median co-located. For some bacteria, the mean is at the 90th percentile.
On my citizen science site, I kludged in some prediction algorithms which has been well received (including by medical practitioners reporting that it often identifies symptoms that the patient forgot to mention). I would like to improve and validate these prediction algorithms using more traditional methods.
This is a bit of a wandering post as I explore various approaches.
To investigate this, I am using a dataset processed through a single process (Biomesight.com) of 2585 samples (the Population). From these 2585 samples, we compute the percentile ranking of the percentage of each taxa. Of these samples, some 1080 (the Sample) have annotated with self-declared symptoms or conditions with 279 different symptoms. We selected only symptoms with more than 36 samples annotated with that symptom. We will work at the genus level only so each variable is conceptually reasonably independent.
With this data, we will try to construct and then test models for the hundred of symptoms sets available.
Foundations
We start with the t-scores we obtained from regression on the count per million of each genus against symptoms [1 below]. There are multiple of other possibilities as shown below. These were initially explored and none showed significant better results than [1] after running 48,000 models.
- Based on the average, standard deviation, ratio between with and without symptom of the percentage of the microbiome — ignoring not reported [C]
- Based on the average, standard deviation, ratio between with and without symptom of the percentage of the microbiome deeming not reported to be a zero [CN]
- Based on the average, standard deviation, ratio between with and without symptom of the percentile over all samples — ignoring not reported [P]
- Based on the average , standard deviation, ratio between with and without symptom of the percentile over all samples — deeming not reported to be a zero [PN]
- Based on those above or below the Nth percentile between with and without symptom — ignoring not reported [#]
- Based on those above or below the Nth percentile between with and without symptom — deeming not reported to be a zero [#N]
Percentiles can be useful because it transforms the data into a uniform continuous distribution and should always be explored with microbiome data. I will return to this in subsequent posts.
T-scores
Each genus to symptom analysis results in a t-score. We will use the following 4 values in our tests.
- 1.28 (90%)
- 2.33 (99%)
- 3.10 (99.9%)
- 3.73 (99.99%)
In our regression analysis we obtain the following average counts across symptoms. We also factor in prevalence since some significant genus may be rarely seen to determine the expected number of genus that a sample may have reported that matches our regression pattern.
- t-score: 1.28: average number of genus: 317, with prevalence factored in: 94
- t-score: 2.22: average number of genus: 294, with prevalence factored in: 85
- t-score: 3.1: average number of genus: 270, with prevalence factored in: 76
- t-score: 3.73: average number of genus: 243, with prevalence factored in: 64
Number of Standard Deviations
The process of doing +/- 1 standard deviation will eliminate some significant genus, if the value is below zero or above the population (a nominal 1,000,000) the using it as a test becomes moot.
- t-score: 1.28: with prevalence factored in: 94;1 std dev is 38 , 2 std dev is 34
- t-score: 2.22: with prevalence factored in: 85; 1 std dev is 34, 2 std dev is 30
- t-score: 3.1: with prevalence factored in: 76; 1 std dev is 30, 2 std dev is 26
- t-score: 3.73:with prevalence factored in: 64; 1 std dev is with 25, 2 std dev is 22
With 1 std dev, the expected number of match per sample is 16% (thus 4 to 5.4). These numbers are on the edge for usability with Pearson’s Chi Square (for discussion see chi-square test of independence rule of thumb: n > 5). Increasing belong one std dev drops the expected value uncomfortably low.
The chart below shows the number of genus to check against and the red line being 16% (the number of matches expected by randomness). We compute the genus by factoring in the prevalence of each genus. Roughly 30% falls below the magic threshold of having an expected mean of 5.

From this list we will take two apparently independent symptoms at the high end to see how they behave:
- Official Diagnosis: Attention deficit hyperactivity disorder (ADHD) [#264] – with 54 samples and 118 genus
- Physical: Tonsils removed (TR) [#442] – with 45 samples and 122 genus
- Brain Fog (BF) [#289] – with 339 samples and 122 genus and 60 genus
| Symptom | Without | With |
| ADHD – Average Percentage of Matches to Possible | 2.3 | 11.6 |
| TR – Average Percentage of Matches to Possible | 5.4 | 10.3 |
| BF – Average Percentage of Matches to Possible | 4.7 | 8.4 |
| ADHD – Average number of Matches | 2.8 | 6 |
| TR – Average number of Matches | 2.9 | 6.4 |
| BF – Average number of Matches | 0.7 | 1.4 |
| ADHD – Average number of Possible | 52 | 51 |
| TR – Average number of Possible | 52 | 59 |
| BF – Average number of Possible | 15 | 16 |
| ADHD – Average of individual Chi squares | 5.3 | 2.1 |
| TR – Average of individual Chi squares | 4.4 | 3.2 |
| BF- Average of individual Chi squares | 1.3 | 1.6 |
The numbers above interesting and unexpected. with 50 possible matches and 1 standard deviation, we expect 16% to be matched at random. We observe With being below at 11% and without being in the 2-5% range.
With ADHD we tested to see percentage correct with different percentages as a threshold.
| Percentile Match | With | Without |
| 12 | 48 | 72 |
| 11 | 52 | 90 |
| 10 | 61 | 87 |
| 9 | 62 | 81 |
| 8 | 72 | 77 |
| 7 | 77 | 72 |
| 6 | 79 | 64 |
| 5 | 79 | 57 |
To clarify this table, we will use percentile match of 5. We correctly identify 79% of people with ADHD that have that condition. We only correctly identify 57% of people that are normal and identify 43% of normal people are identify as having ADHD. So, with 100 people with ADHD and 100 without, we end up with 79+43 = 122 people predicted to have ADHD, of which 64.5% are correctly identify. Since the incidence of ADHD is about 5%, then for a random population we end up with 79 +(20 * 43) with ADHD suggested from the microbiome. This mean that only 8% of those identified as having ADHD based on the microbiome actually has it. This is clearly a weak predictive tool when used alone.
Ah, Dangerous Assumptions!
The 16% of possible tests is based on the assumption that we have independence between genus. This is a naïve and statistically dangerous assumption. Any one that is familiar with the KEGG: Kyoto Encyclopedia of Genes and Genomes knows that bacteria are far from independent. One feeds the next genus and may also produce toxins that inhibits other genus. If you make the small philosophical step that the mixture of compounds and enzymes plays a major role with symptoms then we walk into a world full of different genus sets producing similar mixtures. We are not in a one bacteria causes a condition world.
We cannot safely apply standard statistical models.
What is the bottom line?
The above was done using the following information:
- A genus or other rank is reported to be statistically significant difference of means. We believe this data may come from a different lab sample processing (assuming reasonableness).
- We ignore the amount of difference. The amount of difference is very dependent on a lab’s processing.
- We have the average and standard deviation from the lab that the sample was processed with.
- Counting those that exceeds the mean +/- 1 standard deviation will likely identify which group that the sample will likely belong to. Above we reached 75% accuracy for both groups if we have a small sample to tune it with.
- We may need a significant number of genus or other rank. We cannot focus on a few select bacteria.
Identifying the my preferred “sweet point” where the probability is equal for identifying into each group is shown above — but requires a significant number of annotated samples for each groups. There may be another novel way…. that the exploration in my next post.
Using Published Studies
Most published studies use very small sample sizes for both control and condition. A Condition-Taxon listing is available here. The number of potential taxon matches (adjusted for prevalence in Biomesight samples) is below.
| Metabolic Syndrome | 48.9 |
| Mood Disorders | 46.0 |
| Type 2 Diabetes | 42.9 |
| Crohn’s Disease | 40.7 |
| Autism | 37.7 |
| Depression | 37.4 |
| Liver Cirrhosis | 36.8 |
| Long COVID | 34.3 |
| Ulcerative colitis | 30.7 |
| Obesity | 28.6 |
| Schizophrenia | 26.5 |
| COVID-19 | 25.5 |
| rheumatoid arthritis (RA),Spondyloarthritis (SpA) | 25.3 |
| Carcinoma | 22.5 |
| Multiple Sclerosis | 22.2 |
| Parkinson’s Disease | 20.7 |
| Inflammatory Bowel Disease | 20.6 |
| hypertension (High Blood Pressure | 20.4 |
| Alzheimer’s disease | 20.1 |
Which bacteria produces ….
A frequent question is shown below
The process is simple, but partially hidden.
First login and then change display to Advance

A new menu will appear
Click Compounds x ( Producers, Consumers). After a little time the page will appear with search option. There are 18,000 items listed.
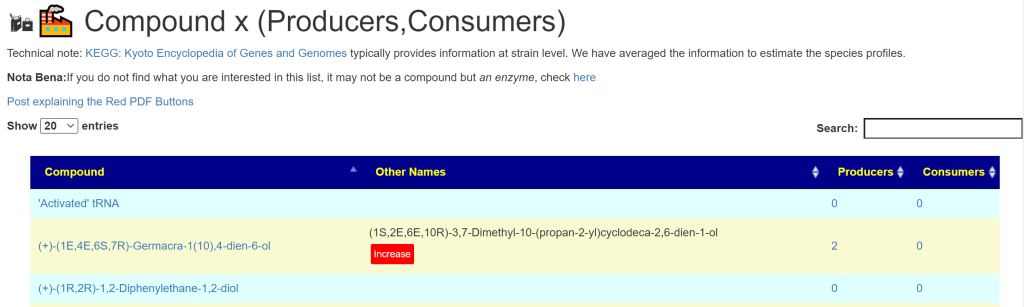
Type the item that you are interest into the search field.
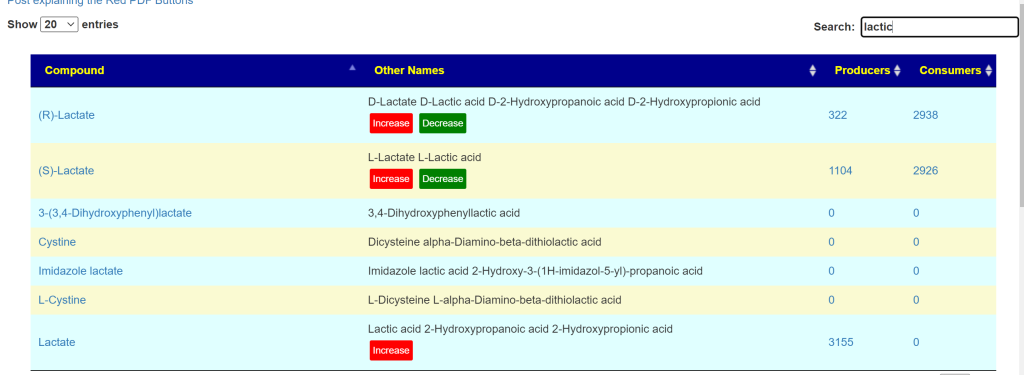
In many cases, this may lead to you needing to read more. Common speech names may not match with the scientific name. In this case we have three forms of lactic acids. the (R)-Lactate is the form that the body has trouble clearing and is associated with neurological issues. There can be some effort required to get up to a reasonable understanding.
What do Zero Mean?
Some chemicals will show 0 0 – this means that it is nether produced nor consumed by bacteria in the microbiome. These may be manufactured or consumed by the body’s cell or obtained from food.
Clicking the Red or Green buttons will down load a PDF which lists the bacteria
Followed by Take and Avoid lists


REMEMBER: These are all computed from calculation and not verified by clinical studies.
The numbers takes you to a list
Protected: Qualified Cloning of MP Data and Web Site
Technical Note: Yield of Applying Different Statistical Methods
Using the five methods described in Technical Note: The Four Winds of Microbiome Analysis, I ran these method on all of the data on the citizen science site of Microbiome Prescription testing for all symptoms that have been self-reported from users of Ombre Labs and Biomesight retail microbiome tests. The data from each lab was done is insolation (you cannot mix data from different processions flows, see The taxonomy nightmare before Christmas… for how the results from the same FASTQ files are reported by 4 different processing flows).
My criteria for deeming a genus significant was:
- At least one method reported P < 0.01
- At least two methods reported P < 0.05
The 2 @ P < 0.05 is a bit of shooting from the hip; I expect some correlation between methods but not sufficient to have that adjusted P value to be outside of the range 0.0025 and 0.01. The statistics on significant genus found is below. The 2 @ P < 0.05 produce only a small contributions,
| Significant Count | P < .01 | P < .05 |
| 2312 | 0 | 2 |
| 241 | 0 | 3 |
| 5 | 0 | 4 |
| 55576 | 1 | 1 |
| 17714 | 1 | 2 |
| 10835 | 1 | 3 |
| 6280 | 1 | 4 |
| 711 | 1 | 5 |
| 28660 | 2 | 2 |
| 4361 | 2 | 3 |
| 12876 | 2 | 4 |
| 3646 | 2 | 5 |
| 13523 | 3 | 3 |
| 6636 | 3 | 4 |
| 4700 | 3 | 5 |
| 49501 | 4 | 4 |
| 9004 | 4 | 5 |
| 29060 | 5 | 5 |
326 Symptoms Had significant Statistical Associations
An example of symptoms with a number of associations is shown below. You may examine them here Citizen Science Symptoms To Genus Special Studies.
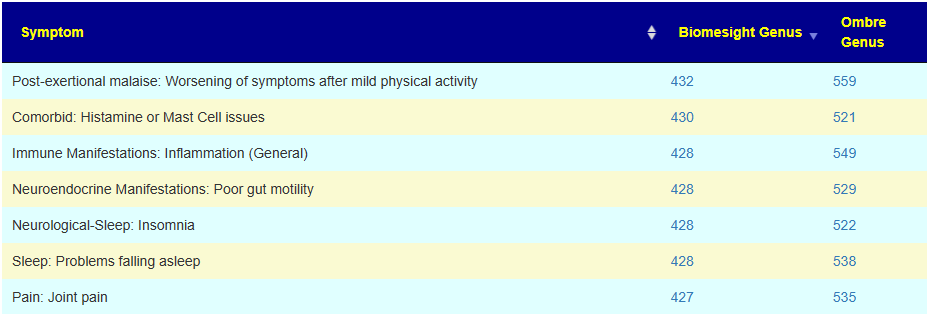
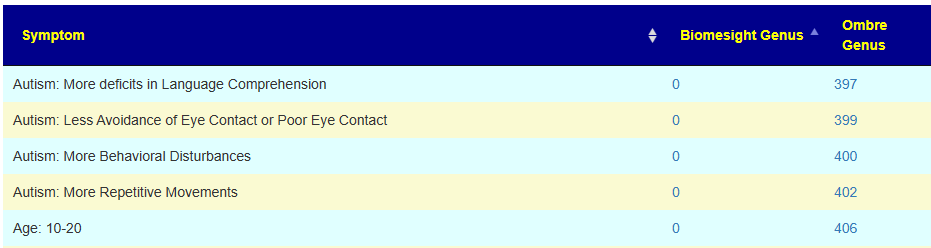
There are some great contrasts between these two labs

With the next set being very interesting. I know that there is a significant subset of 0-20 years old autistic children whose parent have done their microbiome and uploaded. This appears to be reflected in the data. It does call into questions the 10-20 yo associations because of the likely over-representation of autism in this range group.
All of this data is freely available at:
Drilling Down to Genus Involved
If on the above page you click on the count, you will be taken to a sortable table showing the genus. In the example below, we look at the most significant (i.e. P < 0.01 for all five methods).
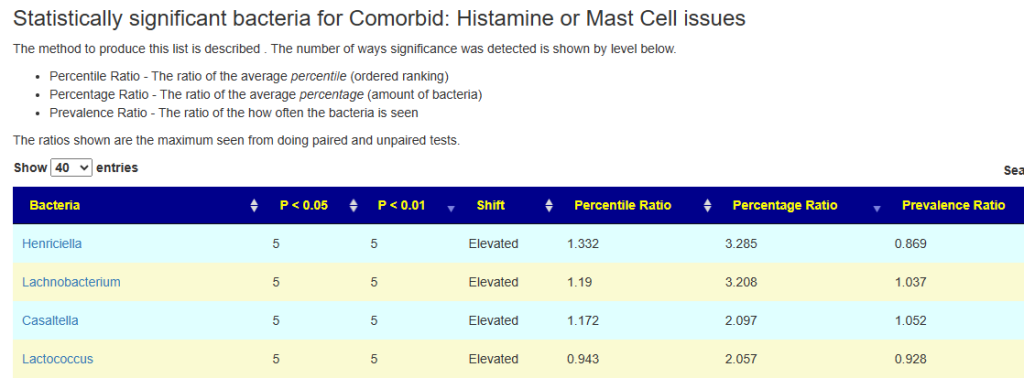
Looking at Lachnobacterium, we see the expected pattern
- The odds of seeing this genus for people with mast cell issues in slightly elevated (1.037)
- The percentage seen is 3.2x what the average for others are
- The percentile is about 1.19x higher
Looking at Lactococcus we see a more confusing picture
- The genus is seen less often (0.928)
- The amount seen with this genus is found is actually much higher (2.057)
- The percentile ranking is slightly lower (0.943)
Both Percentage and Percentile numbers are the maximum using paired and unpaired statistics which may partially account for an apparent contradiction.
Bottom Line
The purpose of this post was to illustrate the data produced from using the five different ways of finding statistically significant association of genus to symptoms. For many readers, this data may be difficult to accept because it disagree with the common sense view of the microbiome that they are working with.
For example, if a genus is seen more often, then you would expect the average amount to be higher. This is often false looking at real data. Understanding the microbiome means discarding simple mechanical models and understanding a complex world of interactions with cascading consequences.
Note: Why we have so many association… Sample Size!
For each of these analysis we have over 1000 annotated samples. As sample size increases, the ability to detect significance goes up significantly.
Pending Work
Tuning parameters:
The numbers will be changing as I tune thresholds. At present:
- I raised the number of times that a genus need to be reported in annotated samples to 36 (i.e. around 0.3% prevelance)
- Number of cases with symptoms reported to 30
This reduced the volume of results (we are not saying less significance — we are filtering by rarer occurrence).
The next step is applying this to an individual microbiome result for a person with one or more symptoms. This means determining which bacteria are the greatest probable contributors and the weight to be given to each for determining a course of microbiome modification.
Technical Note: The Four Winds of Microbiome Analysis
This is one of a continuing set of posts on Microbiome Analysis: Technical Notes on Microbiome Analysis.
Many studies use just one method of analysis: Means of the Counts for bacteria very often seen. The reason is likely conditioning from their education and not knowing how to handle a variety of statistical complexities.
For my analysis I tend to use the following four methods:
- Means of Counts for those reporting this bacteria [Reported]
- Means of Counts with zero for those not reporting [All]
- Prevalence (see Technical Note: Prevalence, Average and Not Reported) [Prevalence]
- Means of Percentiles for those reporting this bacteria [Percentile]
Mini-lessons on the methods
For those folks who may be rusty on technical aspects
Means of Counts for those reporting this bacteria [Reported]
With this method we compute the average and variance measures for each group based on the percentage of each bacteria in the sample, for each sample in the two test groups (with and without brain fog). Not reported values are ignored.
From these numbers we then compute the t-test statistic (see Hypothesis Test for a Difference in Two Population Means ). From this t-test statistic, we lookup or compute the probability of them being the same. If there is less than 1% chance of the two sets being the same, then we say P < 0.01; 5% chance is P < 0.05; 0.1% change is P < 0.001.
Means of Counts with zero for those not reporting [All]
With this method we compute the average and variance measures for each group based on the percentage of each bacteria in the sample, for each sample in the two test groups (with and without brain fog). Not reported values are deemed to be zero.
From these numbers we then compute the t-test statistic (see Hypothesis Test for a Difference in Two Population Means ). From this t-test statistic, we lookup or compute the probability of them being the same. If there is less than 1% chance of the two sets being the same, then we say P < 0.01; 5% chance is P < 0.05; 0.1% change is P < 0.001.
Prevalence (see Technical Note: Prevalence, Average and Not Reported) [Prevalence]
With this method we determine the percentage of time that a bacteria is seen in each group. A simple example would be the incidence of finding salmonella bacteria in people with food poisoning may be 80% and in people without it, 10%.
The method is well known and described here: Comparing Two Independent Population Proportions.
In this case, we obtain a z-score instead of a t-test statistics. From the z-score, we lookup or compute the probability of them being the same. If there is less than 1% chance of the two sets being the same, then we say P < 0.01; 5% chance is P < 0.05; 0.1% change is P < 0.001.
Means of Percentiles for those reporting this bacteria [Percentile]
With this method we compute the average and variance measures for each group based on the percentile of the bacteria in the sample across some reference set. In this case, not reported values are ignored.
Using percentiles is not common in life and physical science, it is used occasionally in economics. The use of percentiles transform the percentages in the first two methods into a uniform distribution. There are other methods — see Transforming Non-Normal Distribution to Normal Distribution.
From the percentile we then compute the t-test statistic (see Hypothesis Test for a Difference in Two Population Means ). From this t-test statistic, we lookup or compute the probability of them being the same. If there is less than 1% chance of the two sets being the same, then we say P < 0.01; 5% chance is P < 0.05; 0.1% change is P < 0.001.
Means of Percentiles for those reporting this bacteria [Percentile] — NOT DONE
With this method we compute the average and variance measures for each group based on the percentile of the bacteria in the sample across some reference set. In this case, reported values are used. In terms of a reference set, if the prevalence is 50% and the reference set only uses reported values, we simply adjust the numbers as follows:
Percentile(Include Null) =Percentile (Not Null)+ (100- Prevalence Percentage)
Using percentiles is not common in life and physical science, it is used occasionally in economics. The use of percentiles transform the percentages in the first two methods into a uniform distribution.
From the percentile we then compute the t-test statistic (see Hypothesis Test for a Difference in Two Population Means ). From this t-test statistic, we lookup or compute the probability of them being the same. If there is less than 1% change of the two sets being the same, then we say P < 0.01; 5% chance is P < 0.05; 0.1% change is P < 0.001.
Note on Including or Excluding Null Values
If you exclude null values, you will often be indirectly including prevalence into the statistics measurement. Including null values, you are indirectly excluding prevalence. IMHO, to get the most amount of information from the data, do both.
Analysis Pattern
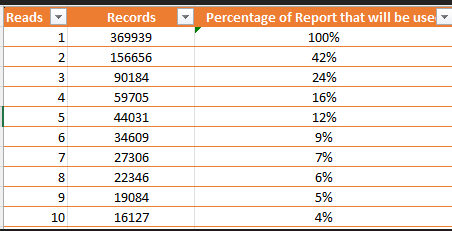
I have a great preference for percentiles because it transforms the VERY non-normal distribution of bacteria into a uniform distribution. One complicating factor with the common 16s tests is the number of reads required to deem a bacteria is there with a reliable measure. Many strains are single reads. If you require more reads, then the number of taxonomy items report drops quickly as shown in the table below.
To give a concrete example with real data, I am using the samples donated to my citizen science site that were processed through the UK based 16s provider, Biomesight.com. I am going to take a subset of those who entered self-reporting symptoms and divide them into two groups:
- Neurocognitive: Brain Fog issues reported (N:328)
- Neurocognitive: Brain Fog issues not reported (N:700)
The high number of Brain Fog is likely a byproduct of Long Covid in the population with those people willing to beat the bushes to find answers.
I am going to look only at genus level for illustration. A prior analysis found that species significance was more pronounced. I attached the data summary below. You can also download the data from Microbiome Prescription Citizen Science Data Repository and do your own data grinding. (Examine the data summary to determine the direction of shifts.)
| Probability | All | Reported | Prevalence | Percentile | Concurrent |
| < .01 | 68 | 20 | 5 | 42 | 0 |
| <.05 | 158 | 61 | 23 | 102 | 1 |
| < .10 | 241 | 102 | 36 | 147 | 0 |
Concurrent agreement between Significant Bacteria is quite dramatic as just one bacteria stands out: Oribacterium. If we exclude Prevalence, we find more.
- < .01
- Desulfosporosinus
- Blautia
- Sporolactobacillus
- Gallionella
- Paenisporosarcina
- Planococcus
- Limnobacter
- < .05
- Phascolarctobacterium
- Helicobacter
- Alkalihalobacillus
- Natronincola
- Alkaliphilus
- Oribacterium
- Cerasicoccus
- Salisaeta
- Anaerobranca
- Acetomicrobium
- Erysipelothrix
- Thioalkalivibrio
- Chryseobacterium
- Anaerotruncus
- Desulfurispora
- Lysinibacillus
- Halanaerobium
- Allochromatium
- Oleomonas
- Marinospirillum
- Moorella
- Agrococcus
- < .10
- Alishewanella
- Halomonas
- Ruminobacter
- Mobiluncus
- Leptothrix
- Dehalogenimonas
- Gemella
- Dorea
- Rikenella
- Brenneria
- Adlercreutzia
- Anaeroplasma
- Candidatus
- Endobugula
- Actinotignum
- Methylocella
- Ligilactobacillus
- Bergeyella
- Dickeya
Let us look at < .10 above, we have 641 genus, so .1 * 641 = 64 false positive would be expected. If you went down that path, you ignored that < .1 occurred over THREE measures. Assuming independence of each measure (for simplicity), then we have (0.1)3 or < 0.001; in other words, 0.001 * 641 = 0.6 false positive. Effectively, every genus listed above is statistically significant at < 0.001 for Neurocognitive: Brain Fog.
Some Visual Representations
I tend to use visuals to better understand processes. In this case, contrary to my expectations, we have quite dramatic differences in appearance. The first one looks like a normal distribution and the others definitely not normal distributions.
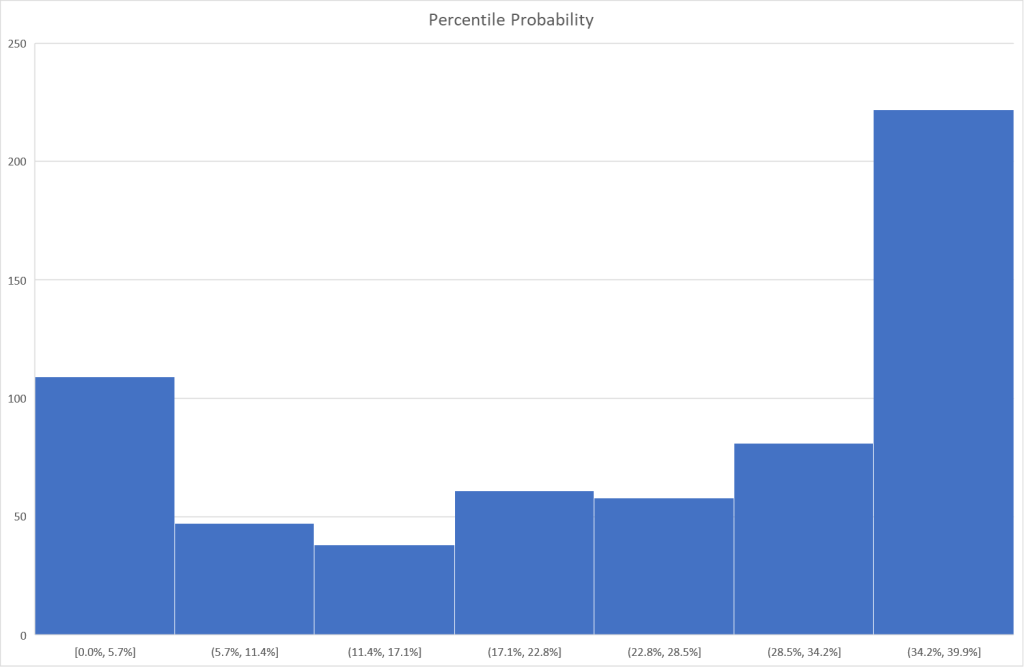

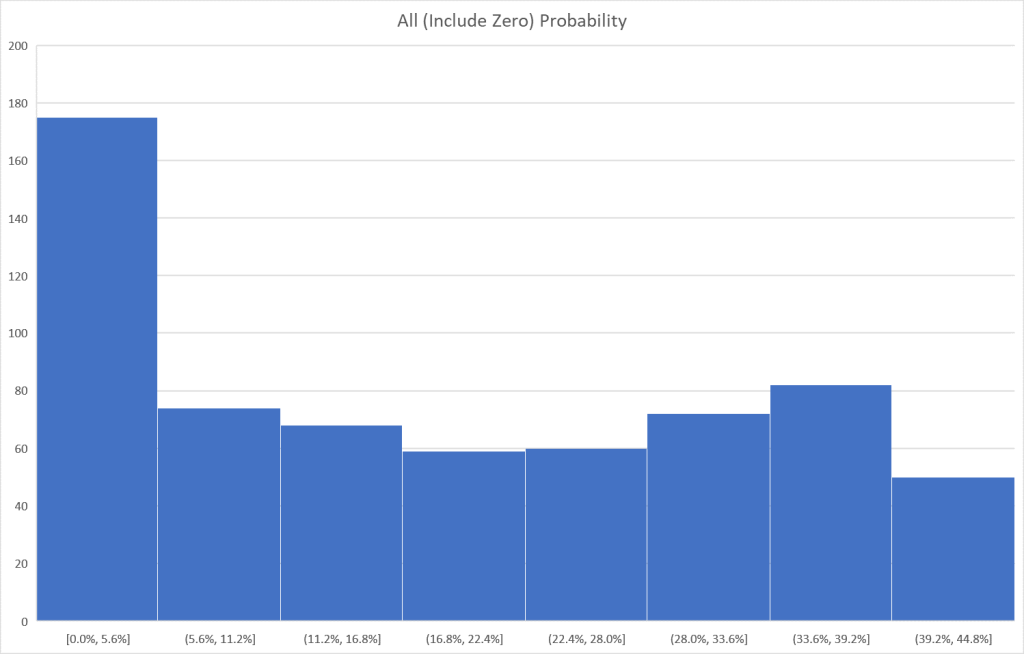
Bottom Line
The purpose of this post was to illustrate that no single method of determining significance is ideal. The use of prevalence is ideal for infrequently seen bacteria but is unlikely to produce results for commonly seen bacteria. It is important to understand the differences and ,in practice, do all five as a pro forma practice for microbiome data analysis.
The next question is simple, how do you treat people with brain fog? My own approach is to obtain a detailed microbiome sample (in this case, biomesight’s) and then identify which genus are sufficiently matching this pattern. From that matching, then use the fuzzy logic expert systems at Microbiome Prescription.com to suggest supplements, probiotics, diets and prescription drugs. There may be alternatice approaches to detemine a treatment approach.
Update: A Statistically Valid Gut Index
I am by training a mathematician, a statistician and operations researcher. For years, I have seen different indices proposed and “Frankly, my dear, I don’t give a damn“. I deem them to be the equivalent of Passing with the Wind. Apologies for the pun.
The problem is that they are using the art of clinical or research experience and NOT the science of mathematics.
Over this last weekend I revisited with more rigor my “percentages of percentiles” approach and found that I could create a An Eubiosis Index for the Microbiome without using Art, just mathematics. Eubiosis means the opposite of dysbiosis.
When you go to my profile, you will see your index and a revised chart below.
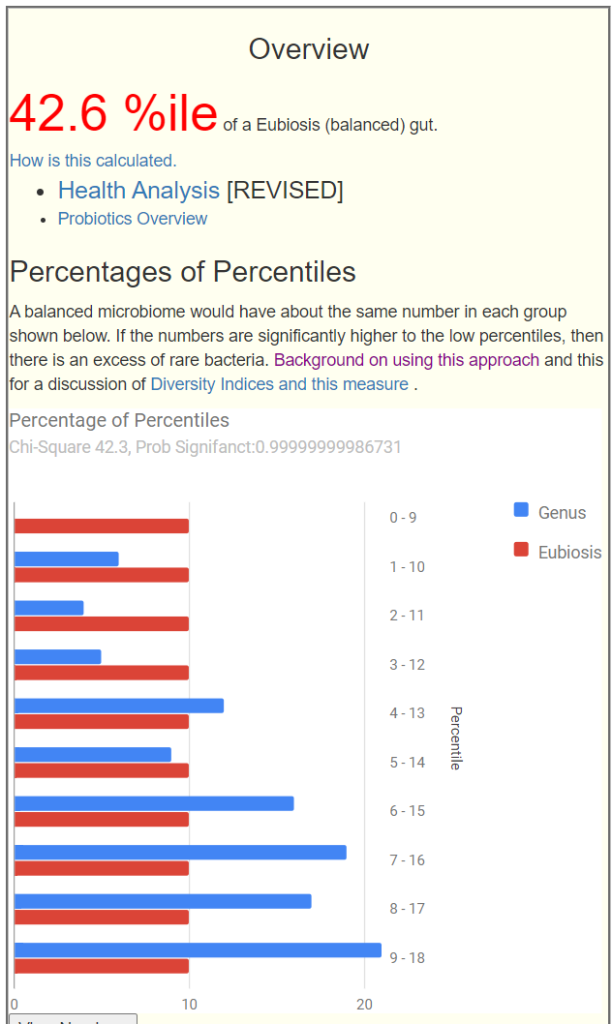
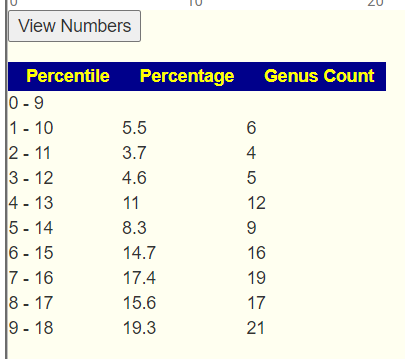
Below the chart, you can click to see the numbers. In case, we see that high percentile dominates.
That’s all folks
If you want technical details see An Eubiosis Index for the Microbiome. The computation is simple and other sites are free to use it with appropriate acknowledgement..
Technical Note: An Eubiosis Index for the Microbiome
Eubiosis is a measure of Representative-ness in the Gut
If you look at a place of employment, ideally you would see each part of society represented in the employees. For example:
- 50% males and 50% females
- 62% white
- 19% Hispanic or Latino
- 12.4% Black or Africian American
- 2% with Autism
- 3% Native American
- .01 with Down Syndrome
- etc
A place that matches (or close to matches) could be said to have 100% Eubiosis – that is the percentage expected in reflected in the employees. This does not reflect this firms profitability or employee turnover rates or any of a dozen of other measures. It is an adjunct measure that is statistically based. Most estimates of gut health are subjective, often based on beliefs or for a specific type of condition. This measure can be low for someone in good health with no symptoms, but the odds are low.
To illustrate this, the following is from one individual over time. Genus and Overall are from special studies.
| Date | Eubiosis | Hawrelak | Genus | Overall |
| 2024-03-05 | 0.7%ile | 12%ile | 30% | 96% |
| 2023-12-06 | 49.7 %ile | 26.1%ile | 100% | 79% |
| 2023-07-03 | 48.3%ile | 16%ile | 79% | 71% |
| 2022-12-28 | 46.2%il | 80%ile | 28% | 99% |
The Eubiosis and Hawrelak both indicates worsening in the latest sample (my usual first question is: Any virus caught between samples? Second question is any vaccinations in the 6 weeks prior to the sample). Overall Citizen Science being high for symptoms indicates a strong pattern match to symptoms — which implies a worsening microbiome.
Below we have a bad Eubiosis, 3.9 — why is it bad because the amounts in each range is very far from the expected (statistically). The blue and red bars should be close to each other.
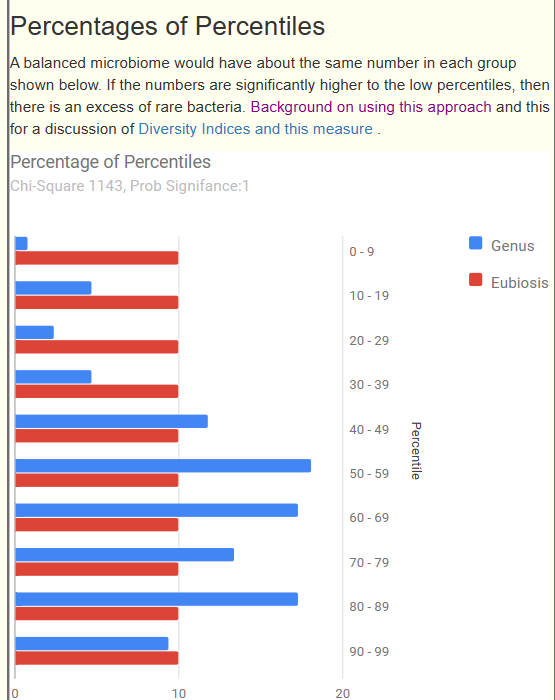
This note in a continuation of an earlier note:
I am a statistician and operations research person by training and experience. I tend to take novel approaches to many issues based on mathematics. These are recorded in this series of notes
I have some 4,600 different unique microbiome samples uploaded to my citizen science site. Most of these are from people with gut issues. Some are from health hackers (i.e. no issues).
A simple Chi2 experiment using percentages of percentiles is done. I bucketized the genus data into 10% percentile ranges resulting in 10 buckets, compute the chi2 and thus we have 9 degree of freedom.
Genus has adequate counts per sample. Species reporting is often very sparse for some tests (depending on the number of reads that the lab set as a threshold, etc.). Genus gives the highest count for a specific taxonomy rank in this dataset.
I then proceeded to plot the values to see what the data looks like. Note the significance levels for 9 degree of freedom below
- 14.7 is 0.1
- 16.9 is 0.05
- 19.02 is 0.025
- 21.7 is 0.01
- 27.9 is 0.001
Over all of the values, we do see some extreme values
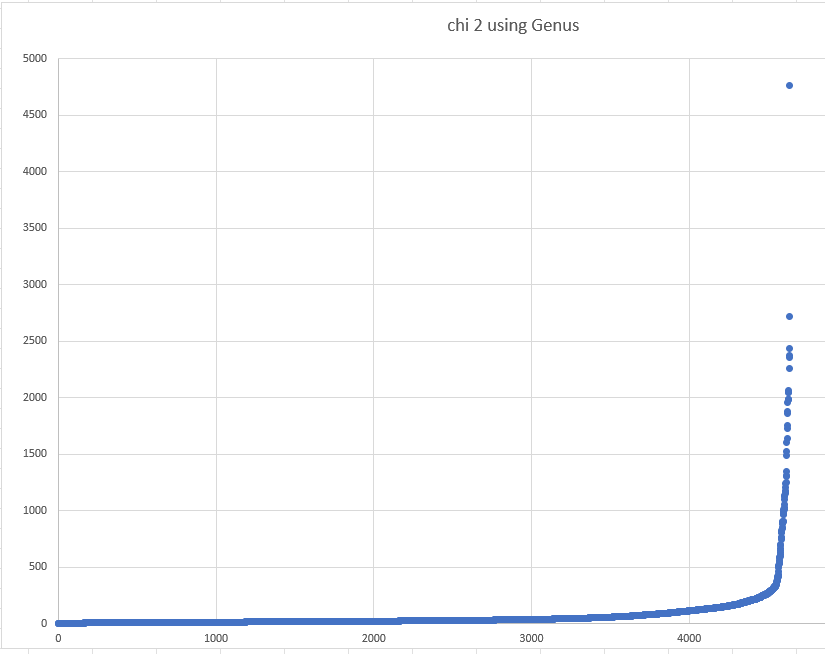
But let us look a less extreme values
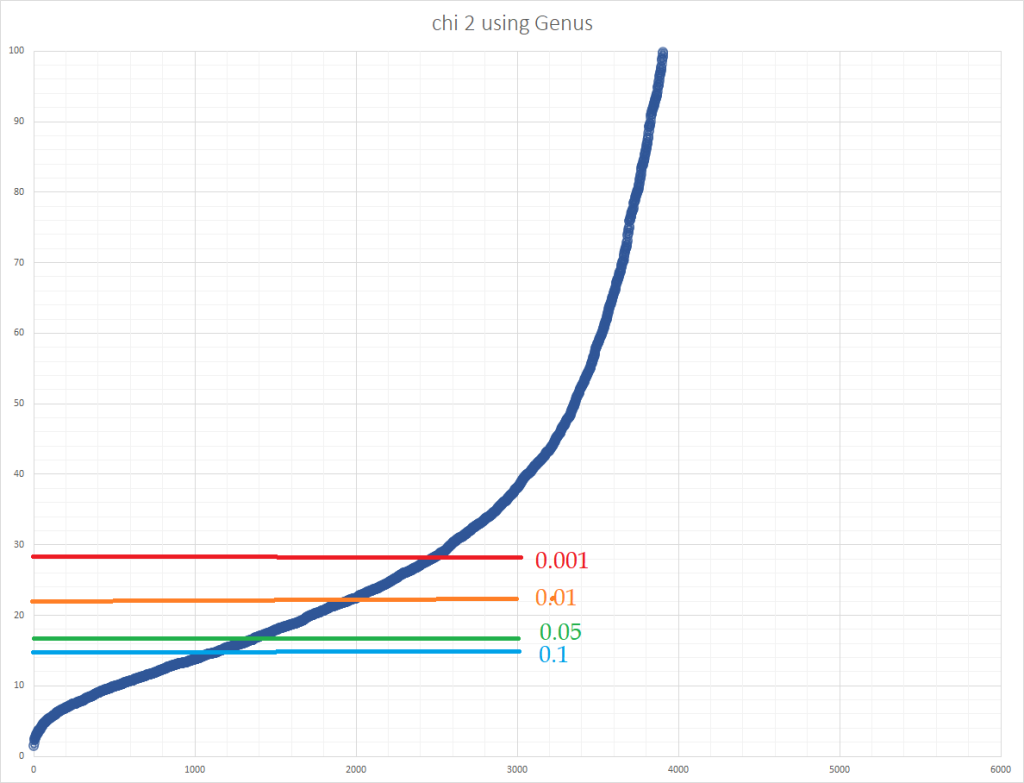
We now need to do a little math assuming there was no significance, i.e. the numbers were happening random.
- 0.1 (aka 10%) means that 90% of the samples would be expected to have a chi2 value of 14.7 or less. We have 1130/4600 or 24% of the samples
We could start working to lower values, but using 14.7 means that 1/4 of the samples at this value may not have dysbiosis. Taking 0.1 and this ratio, we can estimate that we have around a 97% chance of correctly identifying dysbiosis
General conclusion is that a gut without dysbiosis would have a chi2 value of 15 or below for genus.
The Challenge of Getting a “Health Index” for the Microbiome
Looking at a variety of microbiome testing sites I see a lot of “flying by the seats of their pants” being tossed out. IMHO, these sites are soiling their pants — somewhat appropriate for this business :-).
I believe we can create a statistically valid index that works solely off the numbers and not some idealized concept of what a healthy gut should be. We use the above analysis to create this index.
People like have a percentage number for a healthy gut, then the following is suggested (which is actually a percentile ranking):
- Under 15: 100% good
- Over 15: 100 – (Percentile over those over 15)
For lack of a better name (and keeping with naming practices for indices), I will call this the Lassesen Eubiosis Index with 100% being no apparent sign of dysbiosis and good eubiosis.
From the set of samples used above, I extracted a reference table (which may vary according to the test used and the population used). Since I know that the majority of samples have dysbiosis issues, this is likely a reasonable guideline.
| Eubiosis Index | Chi2 |
| 90 | 18 |
| 80 | 21.3 |
| 70 | 24.5 |
| 60 | 29.1 |
| 50 | 35.6 |
| 40 | 45.2 |
| 30 | 65.3 |
| 20 | 106 |
| 10 | 168 |
| 5 | 248 |
The joy of this approach is that it simple, statistically valid and is taxonomy agnostic. No judgement calls are being made on good or bad bacteria.
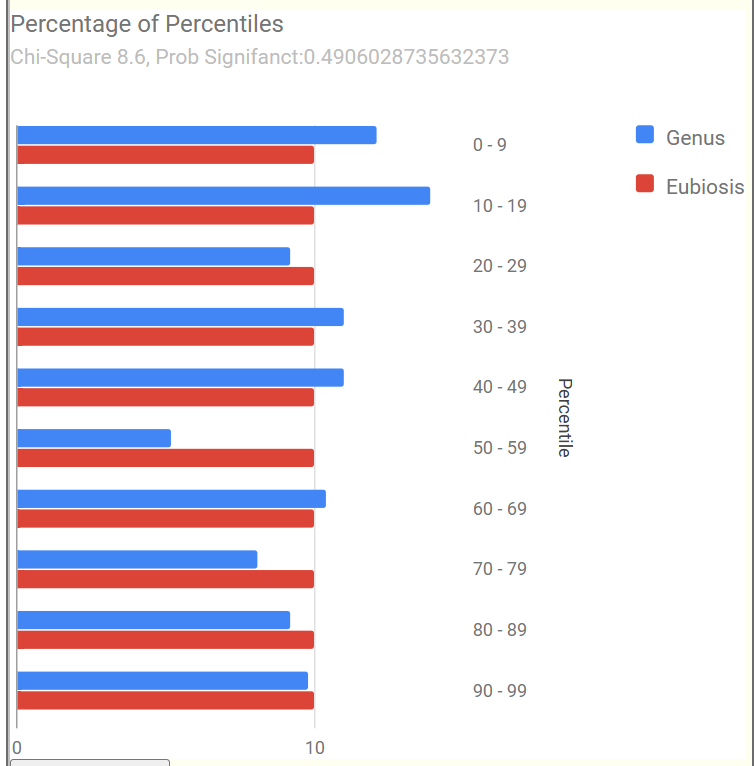
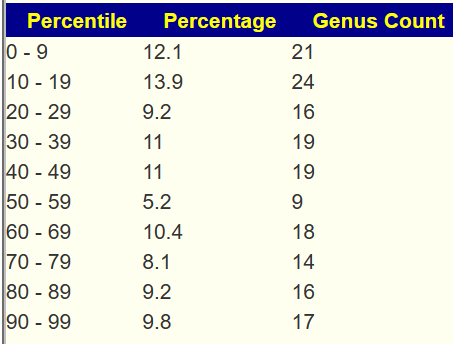
Example of 100% Eubiosis
We see a dip at the 50-59%ile range but this minor disturbance does not register as a likely dysbiosis.
Recent Comments