One thing about dealing with the microbiome is the need to constantly check and test your assumptions and revised from the results of tests. The series of A new specialized selection of suggestions links shattered several reasonable assumptions that appears to be false for most people, namely:
- A surplus of specific bacteria is not the cause (no statistical significance was found)
- A surplus of specific enzymes is not the cause (no statistical significance was found)
- Abundance or deficiency of certain compounds rarely has any statistical significance.
What keeps showing up as statistically significant are:
- An insufficient number of “good guy” bacteria — I do NOT mean traditional internet-lore good guy bacteria.
- An insufficient number of specific enzymes being produced.
In every study we have had 99% of the items being statistically significant at the 99% level being deficiency, i.e. with a condition, the mean of those with the condition is less than the mean of the reference group. The 1% that show up as too high are usually at relatively low z-score and is a surprising good match to the number expected from the False Detection Rate. In other words, they were false significant.
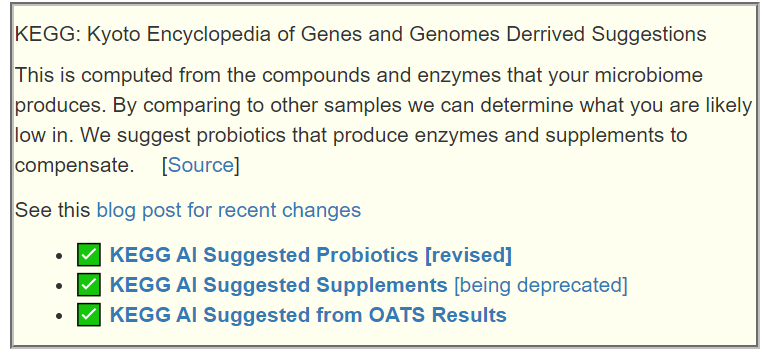
This means that some refactoring is planned. The first one is to give an option to get suggestions ONLY from the undergrowth. The second one, revised the probiotic suggestions from using the genomic data from KEGG: Kyoto Encyclopedia of Genes and Genomes to match the pattern. This has just been done.
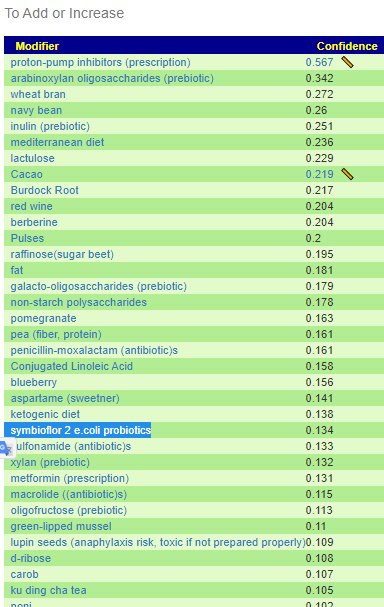
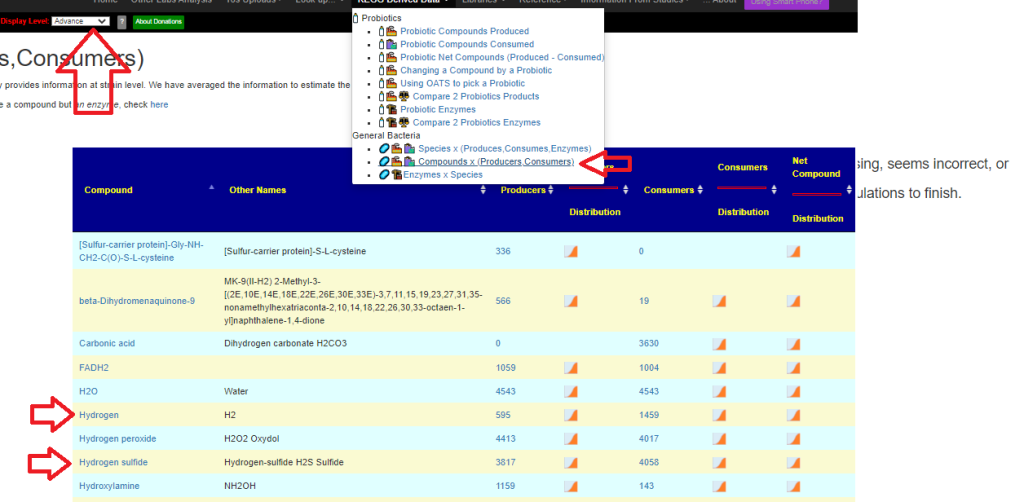
A common pattern that I have seen in the enzyme data has been that the mean of the study group was about 30% of the mean of the reference group. So, the shotgun approach is to flag in a sample all of the enzymes that are less than 30% of the refence group (using lab specific data). From the flagged enzymes, we look at all of the available probiotics as well as potential probiotics species and see which ones produces the most of the missing enzymes. My expectation is that often the best of these probiotics may “kick ass”, in the case of a ME/CFS person “Just started it and wow does this one give me crazy dreams and headaches. I even tried it in the morning and still the same result. Did you just power though or any pointers to deal with the herx”
PLACEHOLDER — VIDEO COMING


Recent Comments