Rotation as a key facet of fixing or keeping a microbiome healthy was first introduced to me in 1999 by Cecile Jadin, MD (Surgeon in South Africa) with the Occult Rickettsia Protocol from the Pasteur Institute of Tropical Disease (where her father worked). I have written about this prior, Rotation and Pulsing: Herbs, Probiotics, Antibiotics [2017], Rotate, Rotate, Rotate and Curcumin [2016], Continuous or Pulse Supplements? [2015]
Some literature on it:
- The effects of antibiotic cycling and mixing on acquisition of antibiotic resistant bacteria in the ICU: A post-hoc individual patient analysis of a prospective cluster-randomized crossover study [2022]
- Optimising efficacy of antibiotics against systemic infection by varying dosage quantities and times. [2020
- The effects of antibiotic cycling and mixing on antibiotic resistance in intensive care units: a cluster-randomised crossover trial. [2018]
- Fighting antibiotic resistance in the intensive care unit using antibiotics[2015].
- “with a 28-day on-off regimen with tobramycin or aztreonam” [2015]
- Antibiotic rotation strategies to reduce antimicrobial resistance in Gram-negative bacteria in European intensive care units: study protocol for a cluster-randomized crossover controlled trial [2014].
- Antibiotic rotation for febrile neutropenic patients with hematological malignancies: clinical significance of antibiotic heterogeneity[2013].
- The impact of an antibiotic cycling program on empirical therapy for gram-negative infections. [2006]
The Microbiome is not a Mechanical System
Think in terms of societies. Think about a gang that steals quality goods at a store. The antibiotic may be having a store detective. Issue solved – no more crime! Wrong, the gang will change to a different crime, they will adapt. It will become a continuous battle and escalation by both sides. Many people view the microbiome as a simple mechanical system unfortunately, bacteria continuously mutate (think of COVID mutations). If the antibiotic kills off 99% of the bacteria, the remaining 1% likely has a mutation that allowed it to survive. Give this 1% a few weeks and we are back to old levels with the antibiotics making no difference.
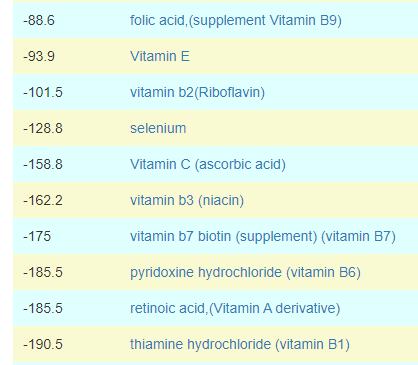
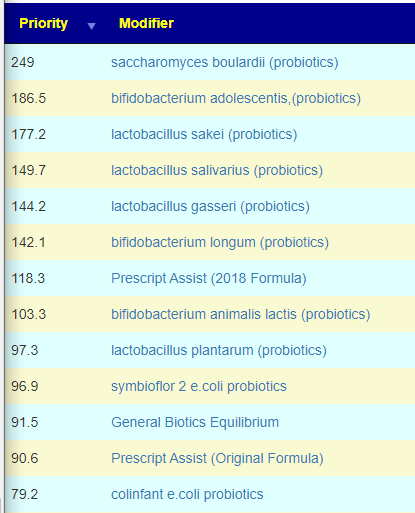
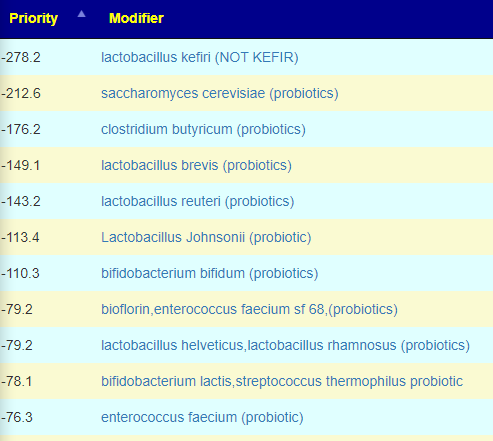
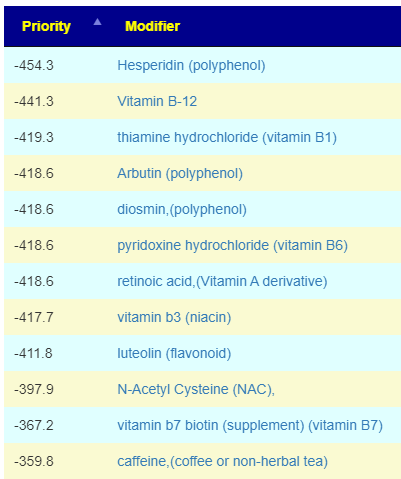
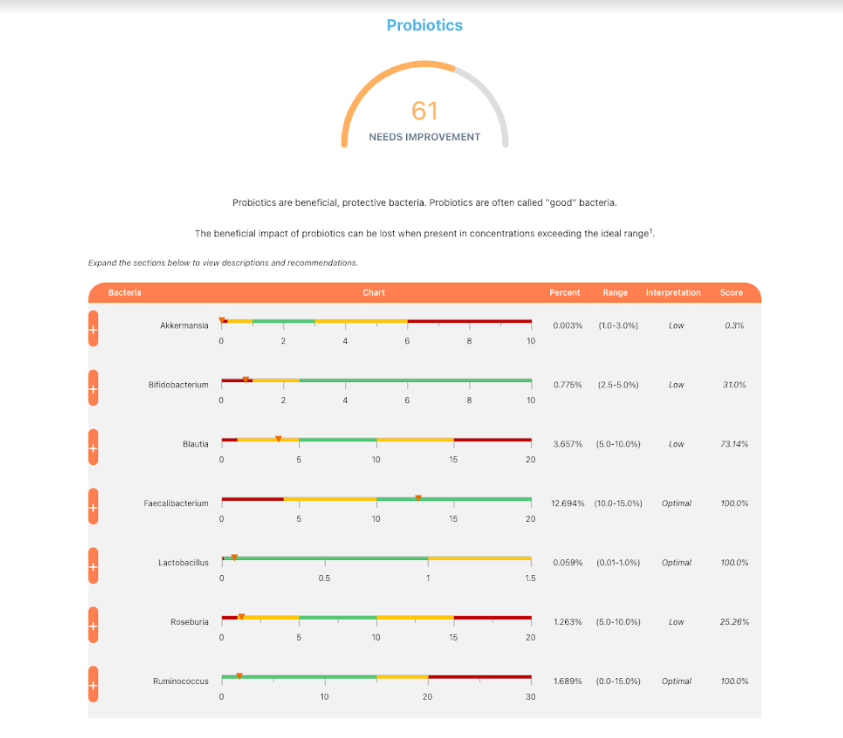
How does this apply to vitamins, amino acids, even food? The simplest explanation is that those items affect the growth of different bacteria. Different bacteria strains produces different compounds, including bacteriocins (natural antibiotics). When you think about probiotics, remember that they often work due to their natural antibiotics (see above for literature).
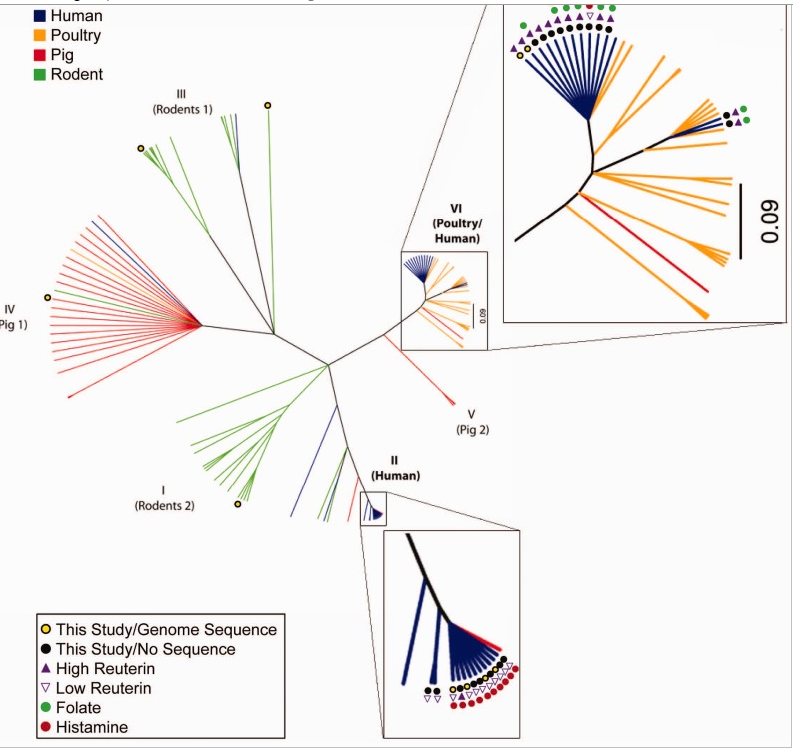
[From prediction to function using evolutionary genomics: human-specific ecotypes of Lactobacillusreuteri have diverse probiotic functions[2014].
To jump to the human body for an example, when you were 20 you likely drank a lots of pop, smoked.. ate a lot of fast food… and were “healthy” and fine. As you body ages (evolved), the same diet would worsen diabetes, weight, and a dozen other conditions that will evolve. The microbiome changes with time, in fact, it changes faster than your body. When you make one set of changes, like running 2 miles a day, the body may act up with structure damage as you age. Professional players in sports do not age well. Going on a specific type of diet may address one issue and trigger a different issue eventually.
What is the answer? All things in moderation. Instead of just one type of exercise, do a variety of appropriate exercises. Instead of a specific diet (which may deliver insufficient minerals or vitamins), rotate around different diets that addresses the issue you are trying to address. Every change you make, changes the microbiome– sometimes in unexpected ways.
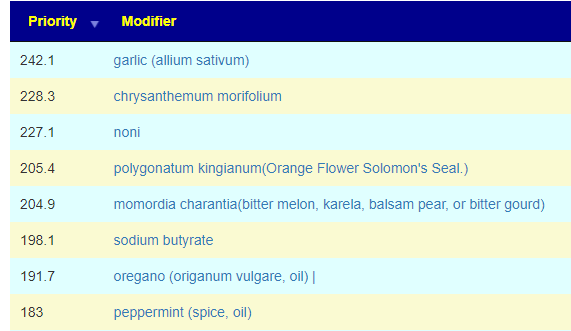
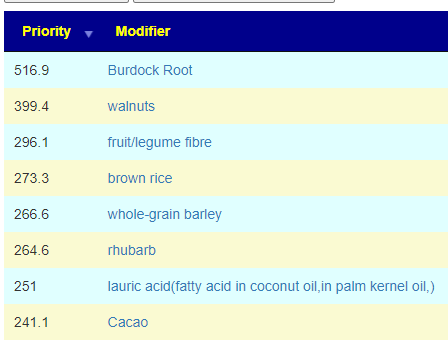
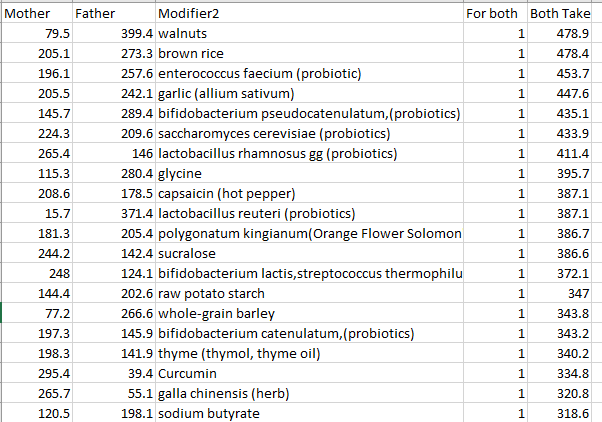
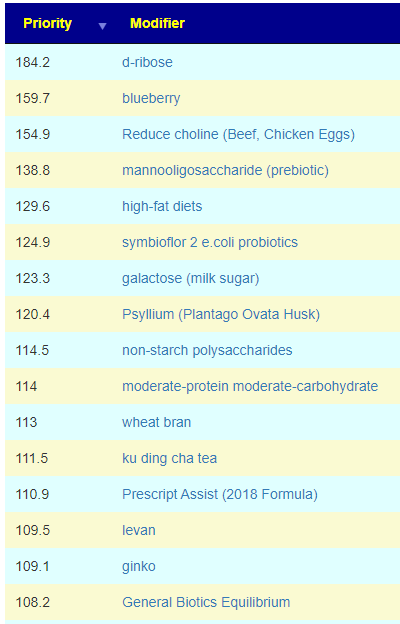
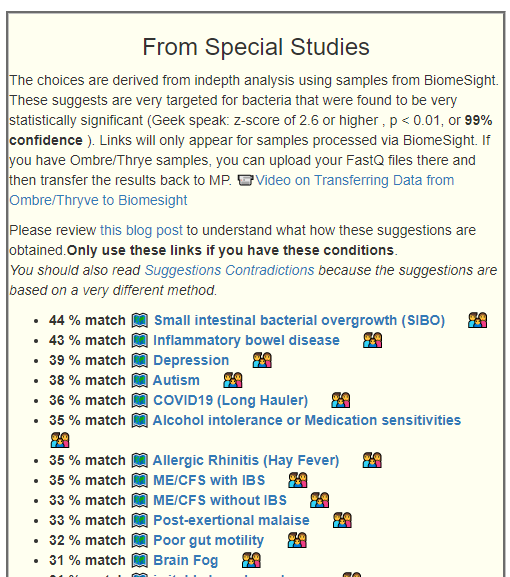
In terms of suggestions from the microbiome prescription site, rotating suggestions is desired — usually there are many suggestions so it should not be hard.
What is the Ideal Rotation?
I do not know, I have inferred from the typical duration of antibiotic prescription that 7-10 days is a reasonable guess. I will give a simple example of one rotation that I do (maintaining), alternating the various forms of gluten (the specific types are listed after each).
- Oats – as porridge: avenin, C-hordeins, γ-hordeins, B-hordeins and D-hordeins
- Rye – as German 100% rye bread: γ-40k-secalins and high-molecular-weight secalins
- Wheat — as typical western baked goods: (ω5-gliadins, ω1,2-gliadins, α-gliadins, γ-gliadins and high- and low-molecular-weight glutenin
- Barley – as porridge: C-hordeins, γ-hordeins, B-hordeins and D-hordeins
My stools will change every cycle – IMHO, there is no perfect stool.
The goal is almost like trying to herd a group of cats.
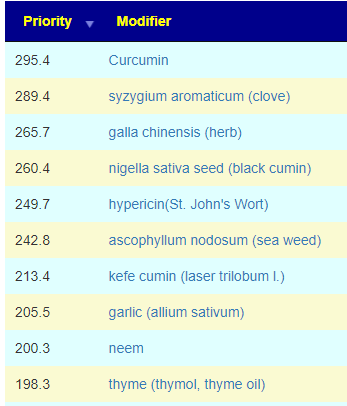
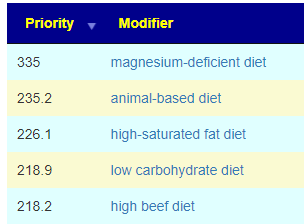
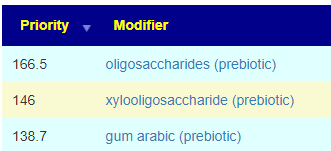



Recent Comments