Conventional clinical studies do not work for Microbiome Prescription. The reason is simple and deals with the model used for those studies:
- Find a cohort with a condition
- Obtain measurement
- Give them one item for n weeks
- Re-measure and publish
This model does not work because our items may be different for each person because they are specific to each person’s microbiome.
Validation Posts
- Cross Validation of AI Suggestions for Nonalcoholic Fatty Liver Disease
- Microbiome Suggestions for Autism — Cross Validation
Alternative Model
For each list of suggestion per person, we see if the substance has been studied for this condition. Examine the results reported in each clinical study. The study may report 80% may have responded, 15% had no response, and 5% has adverse response.
To test with out model, for each person in our cohort of this condition, apply the same process. An ideal result for our cohort would be shown below (subject to sample size adjustments)
- Substance is on the Suggested takes for 80% of the cohort
- Substance is omitted (or very low weight) for 15% of the cohort
- Substance is on the avoid list for 5% of the cohort
If we have a statistical match, then we could conclude that the suggestion process is valid.
This agreement leads to the hypothesis that the cause for being a responder is the microbiome. There has been a few recent studies actually reporting that for some drugs.
Tuning the Artificial Intelligence Fuzzy Logic
Over the last 4+ years, I have been tuning algorithms for both selecting the bacteria to target for suggestions as well as algorithms to get suggestions from the shifts seen in the targeted bacteria. This is an ongoing process. I have focused on a few conditions where I know the literature well and a lot of substances have been tried (one has 9,400 studies on it) and spot checked other conditions (usually, a lot less material to compare to).
My general impression is that each iteration has resulted in more and more suggestions agreeing with clinical studies for the known substances impact on the condition. This general concept of validation is used with many machine learning and artificial intelligence systems, the difference is that we are not validating against our own data but totally independent third party data.
What many microbiome reporting labs may be doing for suggestions
My impression is no-direct-to-retail labs attempt to validate their suggestion beyond a naïve superficial attempt trying to minimize costs.
When a client disclose their diagnosis to a lab, that diagnosis may be matched to a short extract of items that help (using clinical studies from US National Library of Medicine). This is usually done on a one time basis by hiring a nutritionist for a couple of months. Their suggestion may have nothing to do with the actual microbiome report, but because the suggestions work in general — they establish creditability with their clients and drive repeat business.
The second approach is to pick items on a bacteria by bacteria basis. This ignores impact on other bacteria. If they trim their list so that an item is only associated with one bacteria — then they avoid contradictory results in their suggestions, for example, for on bacteria take more fruit, for another bacteria consume less fruit. Again, the suggestions work in general — they establish creditability with their clients and drive repeat business.
My expectation is that labs will provide some evidence in their report — just a single study for each suggestion is sufficient. I see that in many medical gut reports (the studies may be old, but they are given).
A Case Study on one lab’s suggestion
For some labs, I am confused on the basis and their data source.
I went over to the US National Library of Medicine and attempted to find any studies for some of these, because many are not in my list of modifiers. The report identifies why for each one
For the first item checked, Your search was processed without automatic term mapping because it retrieved zero results. And the second item also returned nothing. For the third item, I found one study which reports that appears to report that apple pectin (and thus apple sauce) INCREASES dorea — their suggestion is contrary to the literature.
For the last one, Faecalibacterium, I used my database:
- Lard (I used high fat)
- Chocolate — I found two studies where it INCREASE, not decreases
- Tea — regular tea is reported to increase it, sugar had differencing results
- garlic — no results
- soy — finally agreement with four studies
- lentils — nothing
- asparagus – nothing
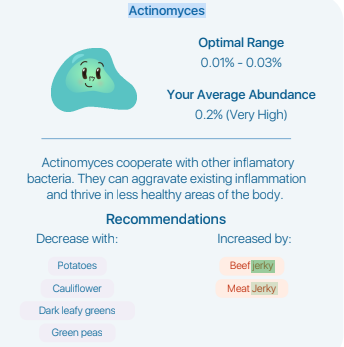
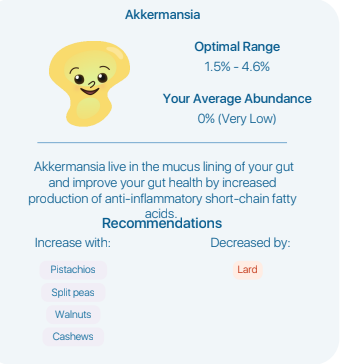
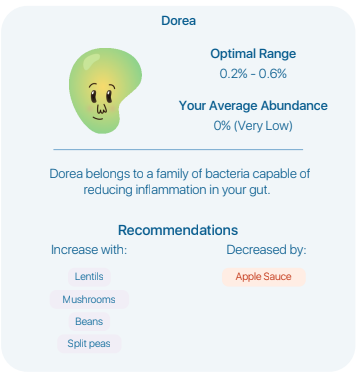

This labs provides zero links to their information sources. I wrote the lab and got this as a response “As for the recommendations, not all recommendations will appear via search as some recommendations are sourced from supplementary materials of publications not indexed by the search. ” I was expecting to get at least one or two links to these mysterious sources back from them… zero.
Recent Comments