This is from a person that have been using Microbiome Prescription for a while with significant success. He messaged me about a new sample and asked me to take a look. It was good timing because I have over the last week refactor forecast symptoms as well as allowing them to be used to generate suggestions. This gave me an opportunity to test the analysis and suggestions.
My impression is that they are both working well with better sensitivity than the older method. Prior posts include:
- A review of a ME/CFS Microbiome Jan 2021
- Evidence of ME/CFS improving using Microbiome Data May 2022
- Comparing 4 ME/CFS Samples with New Tool Jul 2022
- A Microbiome Trek Continues thru the land of ME/CFS. Feb 2023
- Biomesight vs Thorne Tests – Differences Mar 2023
Immediate Back Story
I had covid and an awful toothache a week after.
-PEM
– Reader
-Fatigue on and off specially in the morning. Lowest energy point
-Some brain fog. If I take a binder my brain fog is much better. Guessing that could be related to mold/fungus issues?
-unrefreshing sleep
-hair shedding unless I take Lactoferrin
-difficulty losing weight
Review of Test Results
I am skipping the sample comparison table because the forecast symptoms seem more useful for between samples comparisons.
Percentages of Percentiles
These are helpful overview to see if there are problems. At the top is a chi2 value which a healthy person should have a value below 0.90 . All of his values were .99999…. indicating abnormal shifts.
Prior to 2023
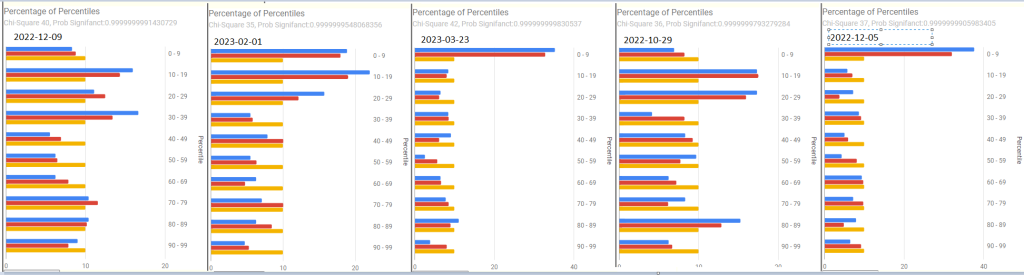
2023, the impact of COVID in the latest sample is quite apparent.
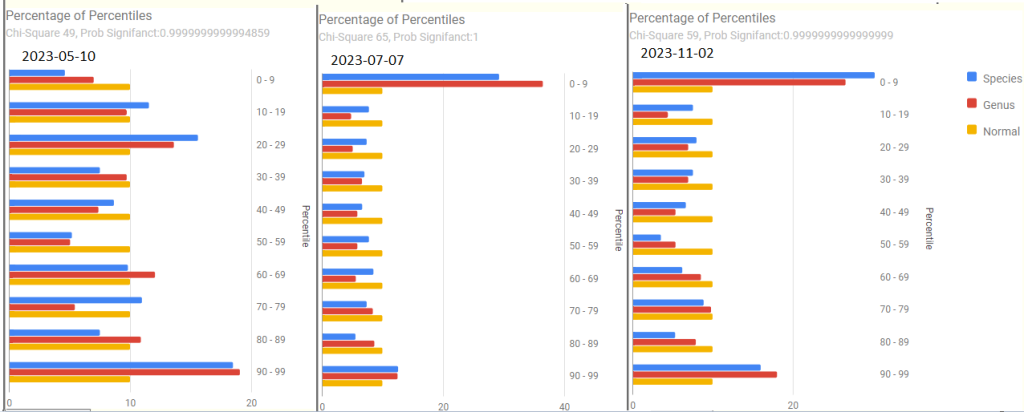
Forecast Symptoms
What shocked (and delighted) me was how accurate the forecasts were, especially with the main issue, PEM being near the top on every sample. The percentage match ebbed and flowed with improvements and set backs (typically from COVID).
- 2023-11-02 – After recent COVID
- 74.2 % match for Neurological: Cognitive/Sensory Overload on 31 taxa
- 63.5 % match for Immune Manifestations: new food sensitivities on 52 taxa
- 61.5 % match for Neuroendocrine Manifestations: cold extremities on 65 taxa
- 60.9 % match for Neurological-Vision: Blurred Vision on 87 taxa
- 59.6 % match for Condition: ME/CFS with IBS on 52 taxa
- 58.3 % match for Post-exertional malaise: Worsening of symptoms after mild physical activity on 36 taxa
- 2023-07-07
- 57.1 % match for Post-exertional malaise: Worsening of symptoms after mild physical activity on 42 taxa
- 54.2 % match for Condition: ME/CFS with IBS on 59 taxa
- 48.1 % match for Post-exertional malaise: Post-exertional malaise on 54 taxa
- 2023-05-10
- 63.2 % match for Post-exertional malaise: Post-exertional malaise on 38 taxa
- 59.3 % match for Post-exertional malaise: Muscle fatigue after mild physical activity on 59 taxa
- 59.1 % match for Pain: Pain or aching in muscles on 44 taxa
- 57.1 % match for Condition: ME/CFS with IBS on 42 taxa
- 2022-12-05
- 82.4 % match for Official Diagnosis: Autoimmune Disease on 51 taxa
- 81.9 % match for Comorbid: Histamine or Mast Cell issues on 83 taxa
- 79.5 % match for Post-exertional malaise: Next-day soreness after everyday activities on 39 taxa
- 71.8 % match for Post-exertional malaise: Physically tired after minimum exercise on 39 taxa
- 2022-10-29
- 64.4 % match for Neurological-Sleep: Inability for deep (delta) sleep on 45 taxa
- 62.7 % match for Official Diagnosis: Mast Cell Dysfunction on 75 taxa
- 60.9 % match for Post-exertional malaise: Next-day soreness after everyday activities on 46 taxa
- 60 % match for Condition: ME/CFS with IBS on 40 taxa
- 55 % match for Post-exertional malaise: Muscle fatigue after mild physical activity on 60 taxa
- 54.5 % match for Post-exertional malaise: Post-exertional malaise on 44 taxa
- 2022-03-23
- 85.4 % match for Neurological-Sleep: Inability for deep (delta) sleep on 41 taxa
- 82.1 % match for Immune Manifestations: Inflammation (General) on 67 taxa
- 77.5 % match for Immune Manifestations: Inflammation of skin, eyes or joints on 111 taxa
- 75 % match for Post-exertional malaise: Worsening of symptoms after mild physical activity on 32 taxa
- 71.4 % match for Post-exertional malaise: Next-day soreness after everyday activities on 42 taxa
Drilling down on PEM
With the revised symptom-taxa association tool, I went to the 2022-12-05 sample because it had very high values for PEM in forecasting. I then went to [Special Studies] and selected all of the PEM items
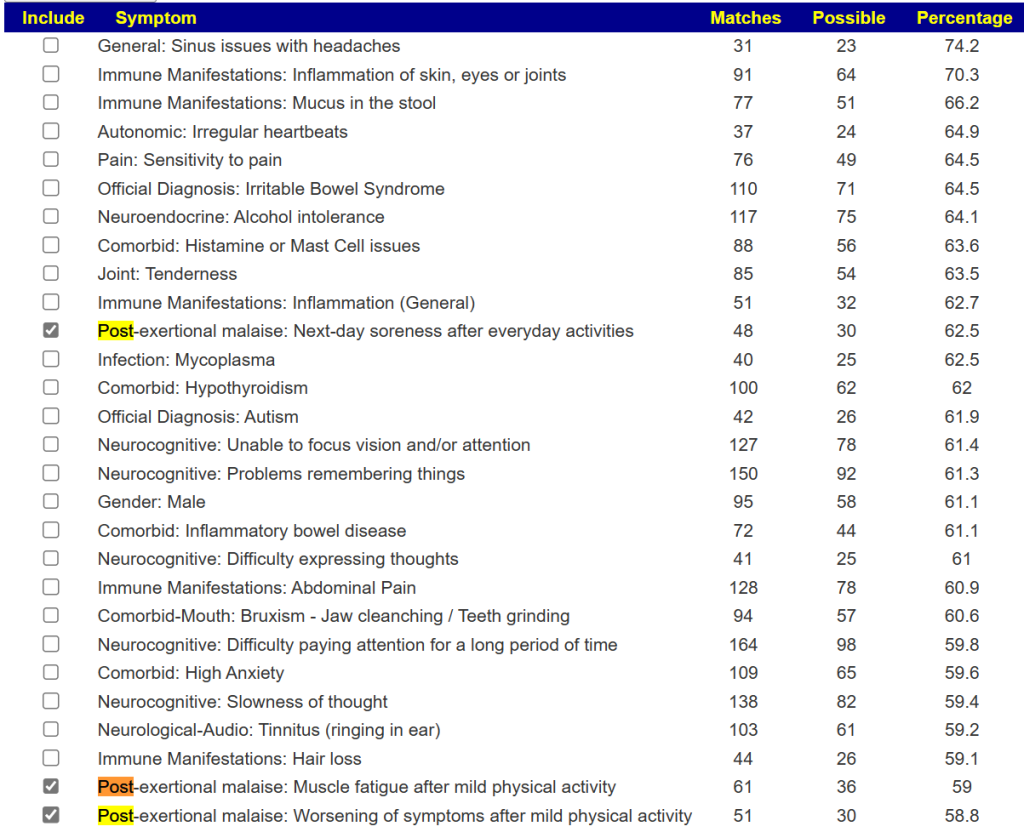
Some 78 bacteria were selected with the following being the top suggestions:
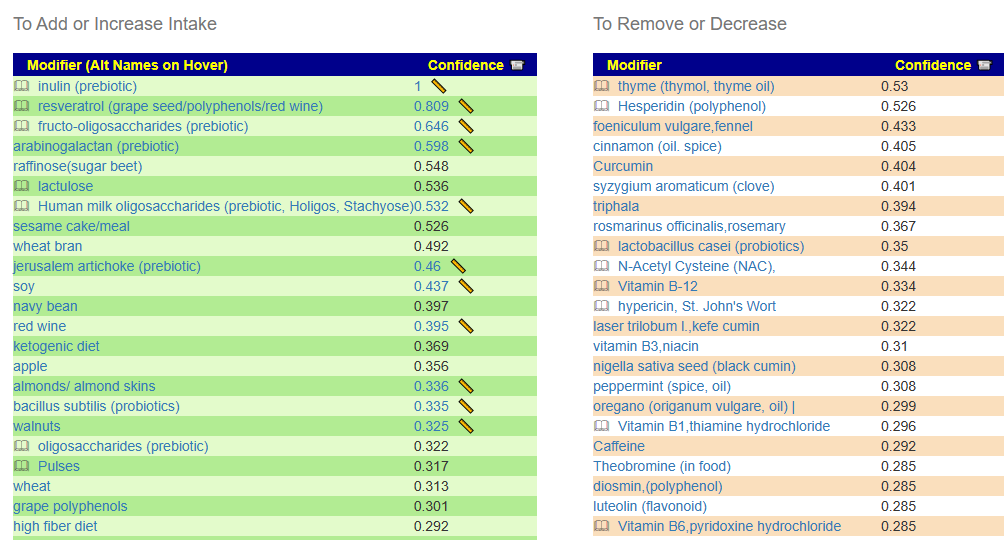
While this is not his current sample, it is reasonable guidance for dealing with the bacteria associated with PEM.
Going Forward
I am going to do [Just give me suggestions] and then special studies selecting only PEM items that are shown. We thus ended up with 5 packages. Looking at the details we have high at 555 -> 277 for high threshold and -542 –> -271 for low threshold
- Antibiotics: the top 2 are used with ME/CFS
- clarithromycin (antibiotic)s[CFS]
- azithromycin,(antibiotic)s[CFS]
- with an IBS antibiotic being close to the top: rifaximin (antibiotic)s
- Amino Acids
- Drugs (OTC etc)
- Flavonoids
- Food
- Herb Spices
- Prebiotics
- Probiotics
- Sugars
- Vitamins
In general, it looks very pro-forma ME/CFS. He may wish to check the PEM suggestions against the details and include anything that is positive.
Bottom Line
I have often used the analogy of going from the port of sickness to the port of health in a sailing ship along a rugged coast. There may be a long series of course corrections needed. While there are still ME/CFS taxa shifts seen in his samples, subjectively the only symptom left of concern is a PEM after playing basketball for a while.
This is a good illustration that taxa/bacteria does not rule — DNA and other factors are involved with symptoms and the ability to tolerate microbiome dysfunctions.
Questions and Answers
Q: I may do another Thorne test if they’re back in stock. Wondering how much aspergillus is holding back healing/gut shifts
- Aspergillus usually impacts lungs and breathing [CDC] (thus ability to get oxygen in). An alternative mechanism for oxygen issues than with hypercoagulation.
Postscript – and Reminder
I am not a licensed medical professional and there are strict laws where I live about “appearing to practice medicine”. I am safe when it is “academic models” and I keep to the language of science, especially statistics. I am not safe when the explanations have possible overtones of advising a patient instead of presenting data to be evaluated by a medical professional before implementing.
I cannot tell people what they should take or not take. I can inform people items that have better odds of improving their microbiome as a results on numeric calculations. I am a trained experienced statistician with appropriate degrees and professional memberships. All suggestions should be reviewed by your medical professional before starting.
The answers above describe my logic and thinking and is not intended to give advice to this person or any one. Always review with your knowledgeable medical professional.










Recent Comments