Backstory
I had no gut problems and only minor health niggles before Covid. I have been hypothyroid for 20 years.
After three rounds of covid (but no vaccines) I have burning and tingling parasthesias in my extremities, neuropathy in my face, plus gut pain, lots of hair loss, and I feel cold a lot and sleep poorly. Some fatigue and shortness of breath, but the other symptoms are much worse.
Trying to increase my bifido and lactobacillus via suggestions on biomesight has barely moved the needle.
All my blood tests work has been normal from a year ago, although now I have a weakly postiive ANA test result.
Analysis
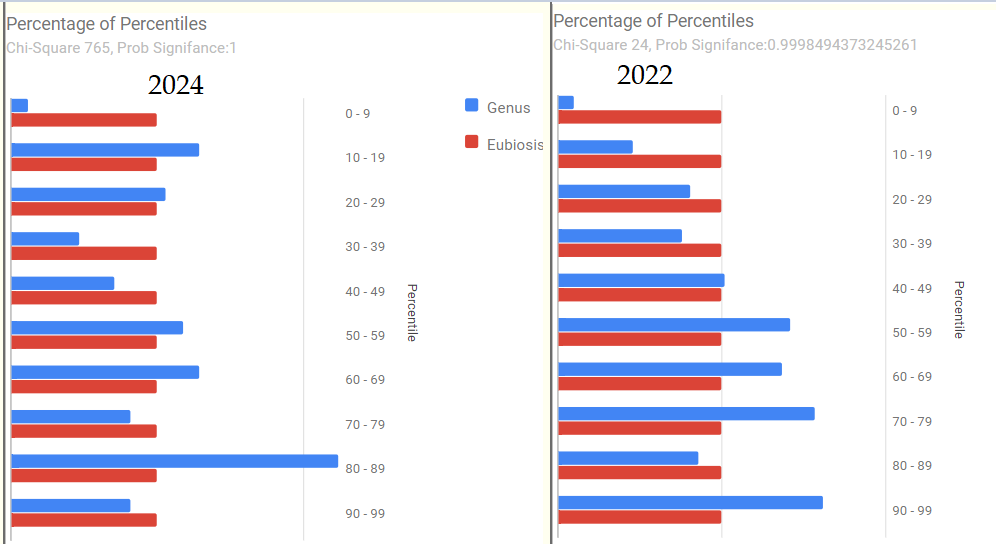
We have 2 samples: 2023-11-20 and 2024-04-26. Both samples are very similar.
High Level Overview
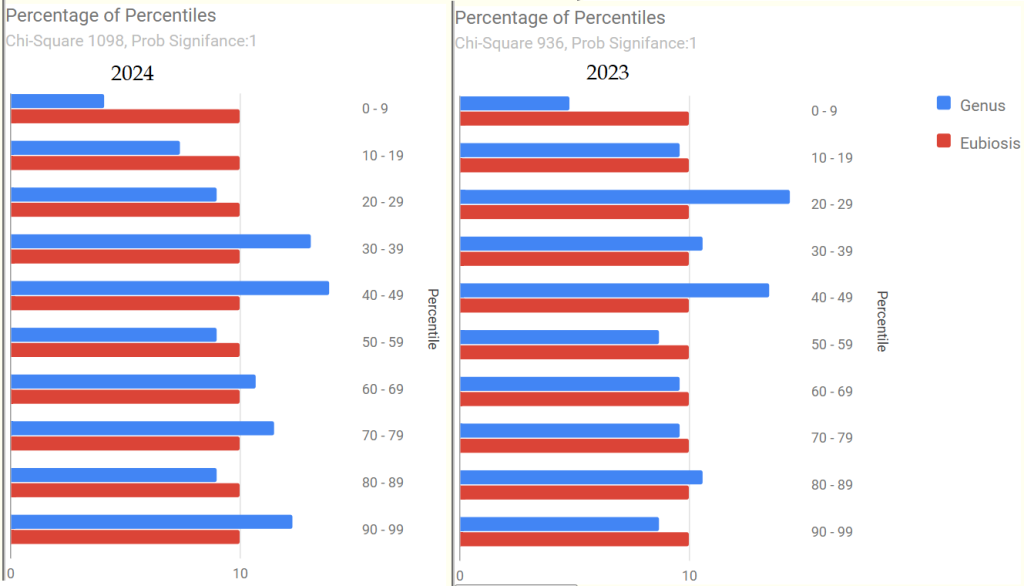
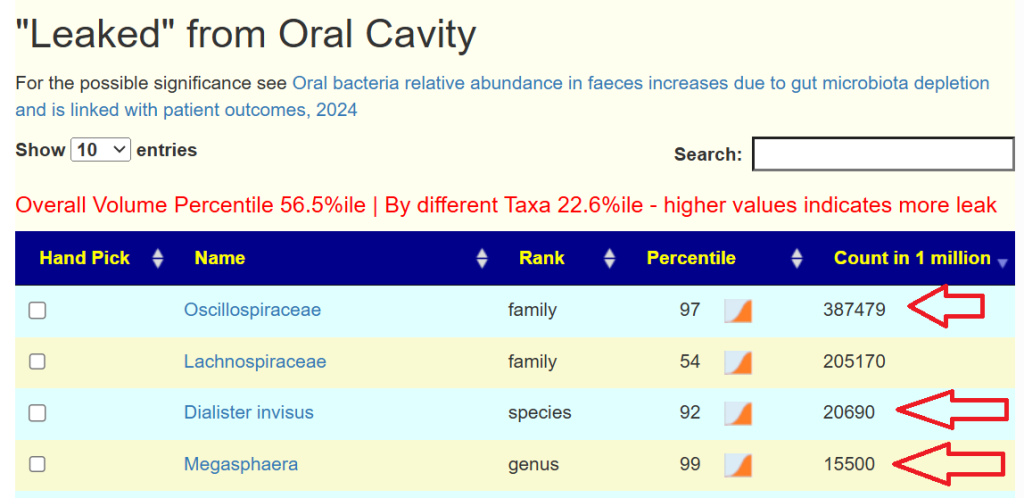
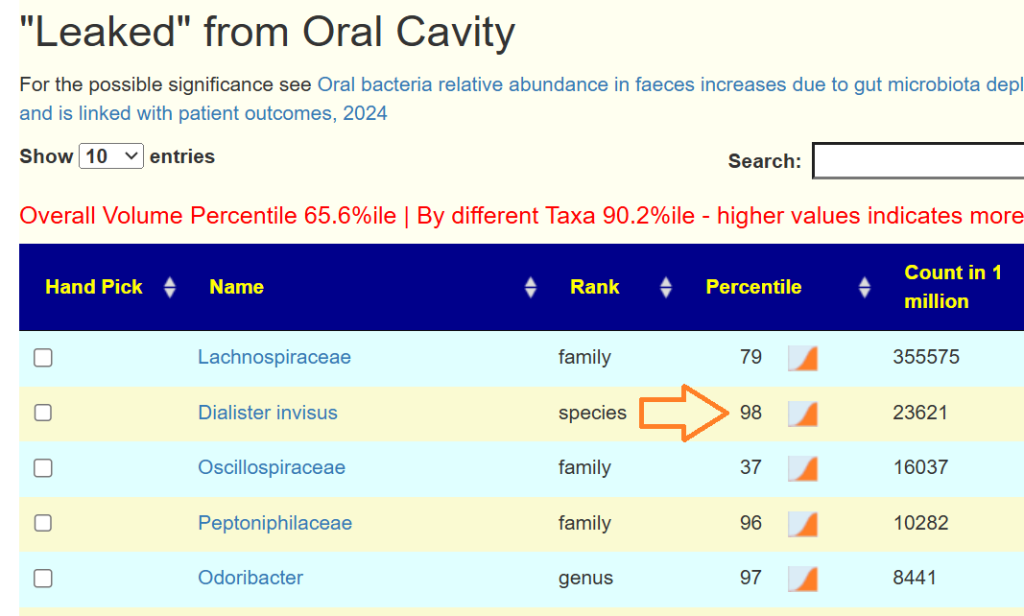
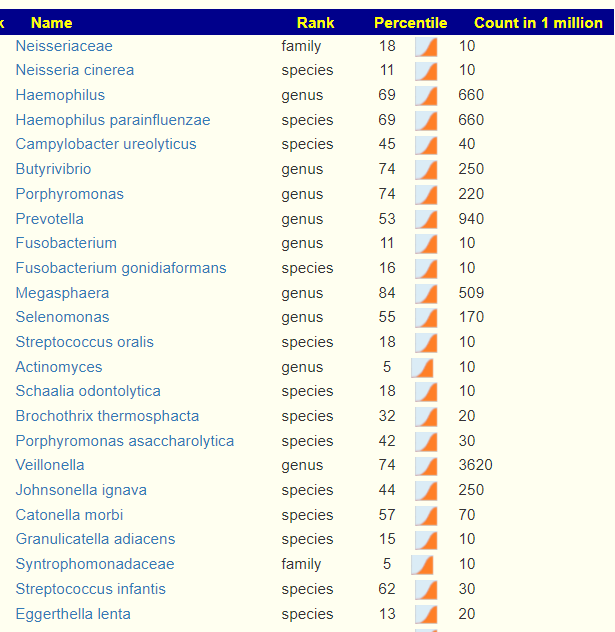
- “Leaked” from Oral Cavity: By volume 77%ile, prior 70%ile
- Dialister invisus 31%ile, 88%ile
- Pedobacter 98%ile, prior 90%ile
- Suggested reading: The Benefits of Probiotics on Oral Health: Systematic Review of the Literature [2023]
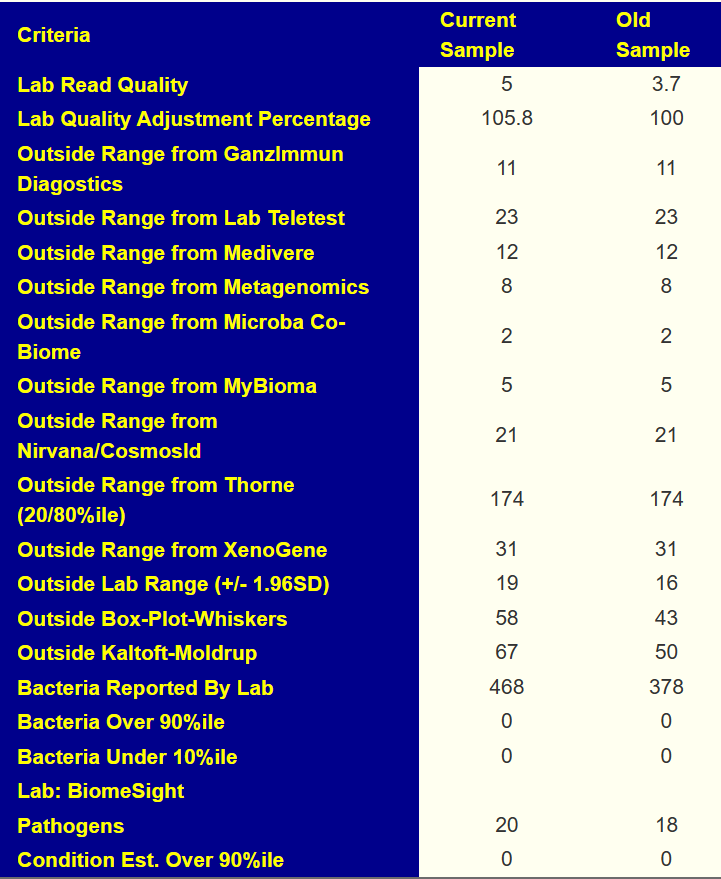
- General Health Predictors: 10 out of range, prior 11 out of range.
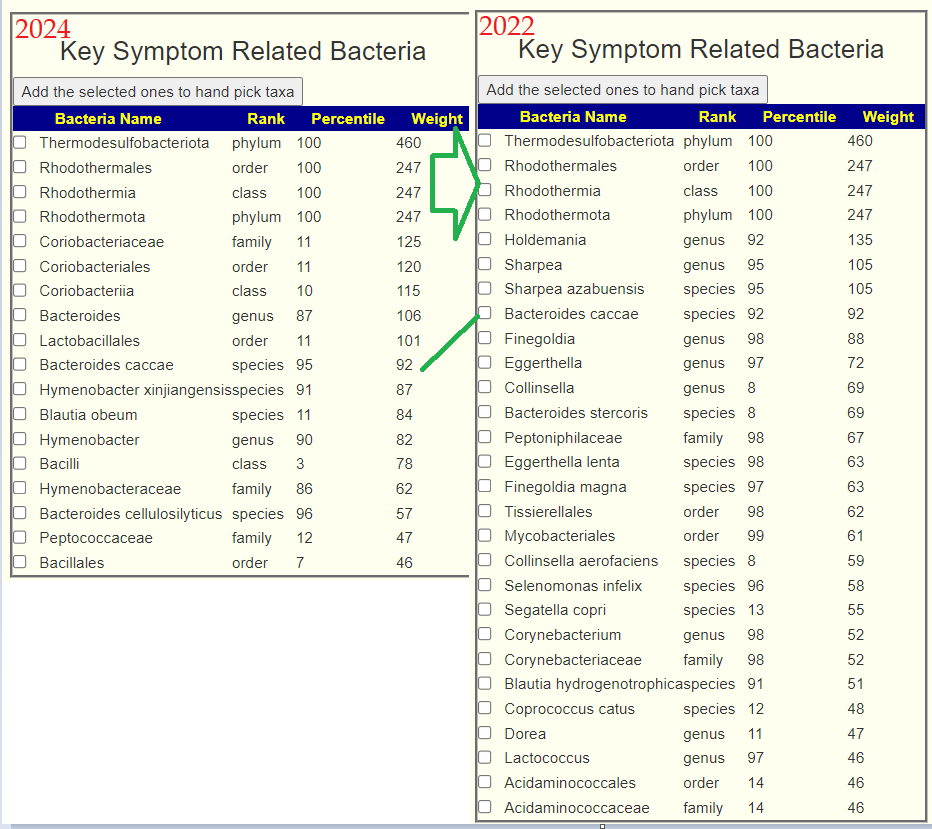
Looking at the “smoking gun”, Pedobacter, we found no known factors to reduce it 🙁
Quick Summary; Past action plan has “barely moved the needle.”
Going Forward
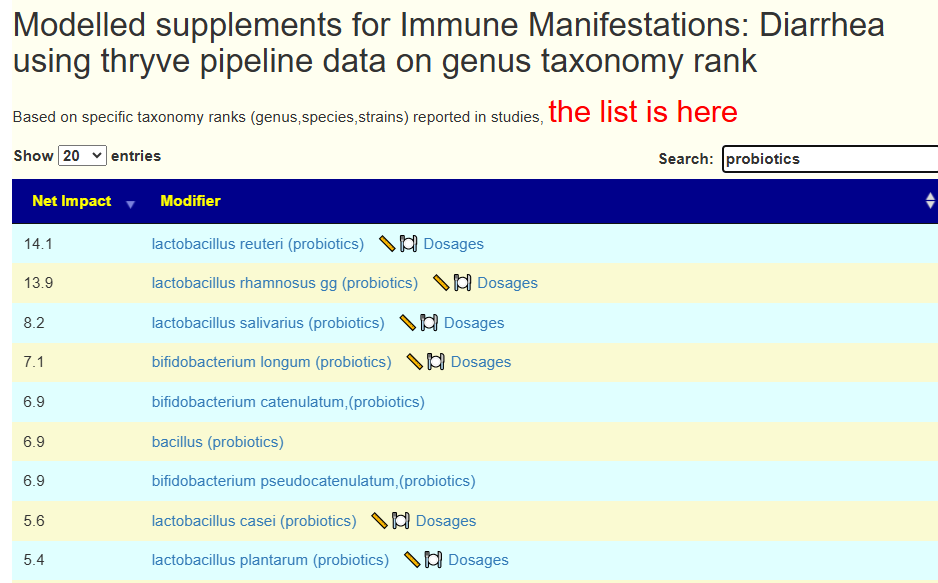
The usual “just give me suggestions” with one addition Bifido and lactobacillus are her primary concern, so I will hand picked those on both samples. and get an emphasis for thoses. We see that one went up and one went down.
| Bacteria | 2024 | 2023 |
| Lactobacillus (only Lactobacillus iners) | 38%ile | NONE |
| Bifidobacterium | 8%ile | 39%ile |
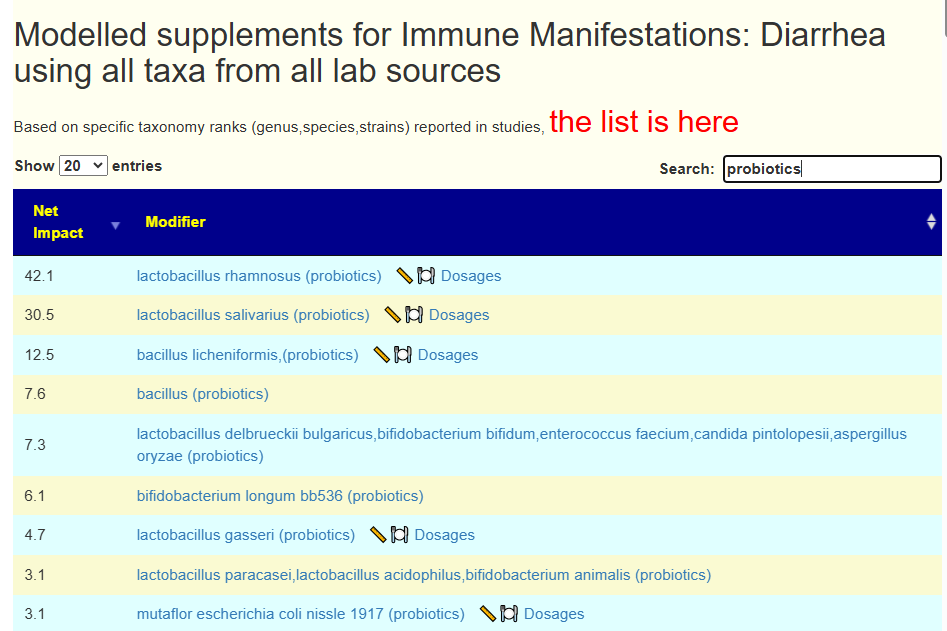
We then do a Uber Consensus with BOTH samples (since they are similar) with a total of 11 sets of suggestions.
- All 11 agrees on these:
- bifidobacterium animalis lactis (probiotics)
- vitamin a
- Olive Oil
- barley
- lactobacillus brevis (probiotics)
- clostridium butyricum (probiotics),Miya,Miyarisan
- ginger and rosemary
- Items to be avoided on all 11 are below
- lactobacillus gasseri (probiotics)
- chitosan,(sugar)
- vegetarians
- blackcurrant
- Dextrin
- Guaiacol (polyphenol)
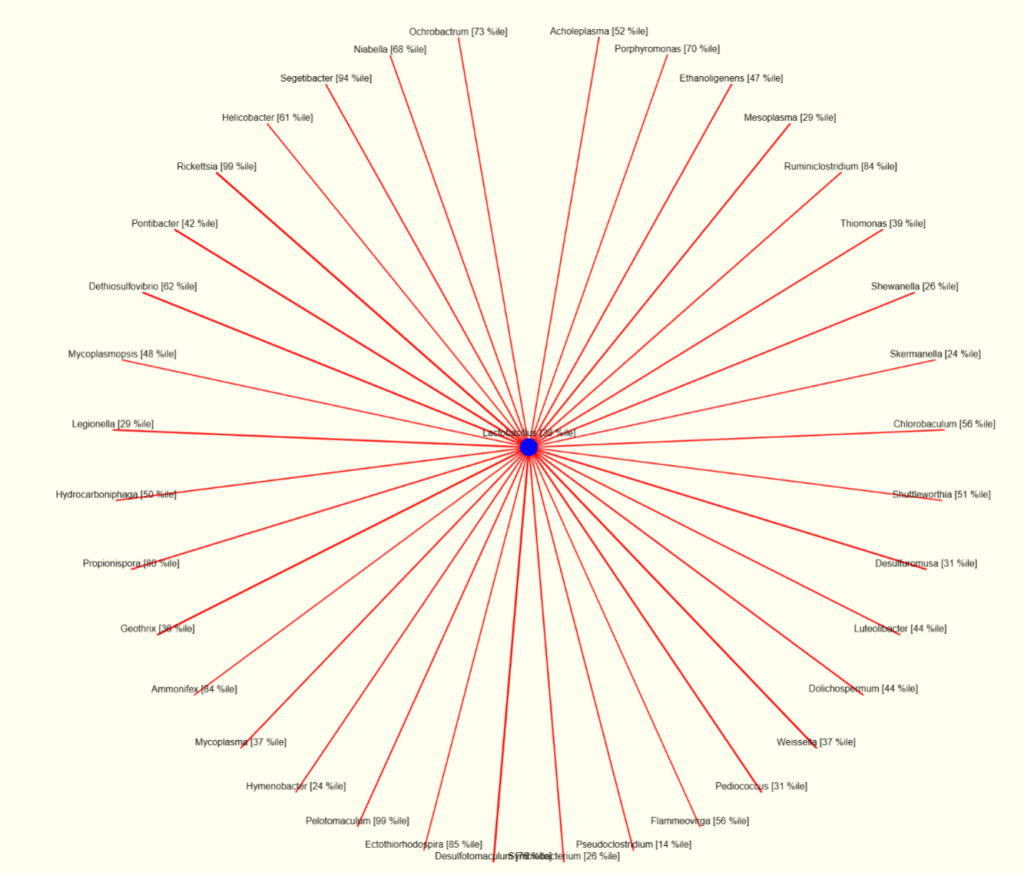
I looked at the interaction charts, the first one is for Lactobacillus and noticed high (99%ile) for Rickettsia and for Heliobacter (61%ile). The last one suggests testing for Helicobacter pylori should be discussed with their MD.
The chart for bifidobacterium shows many of the same bacteria that will surpress bifidobacterium. Note that lactobacillus (top right) is indicated as surpressing bifidobacterium; this agrees with the above numbers.
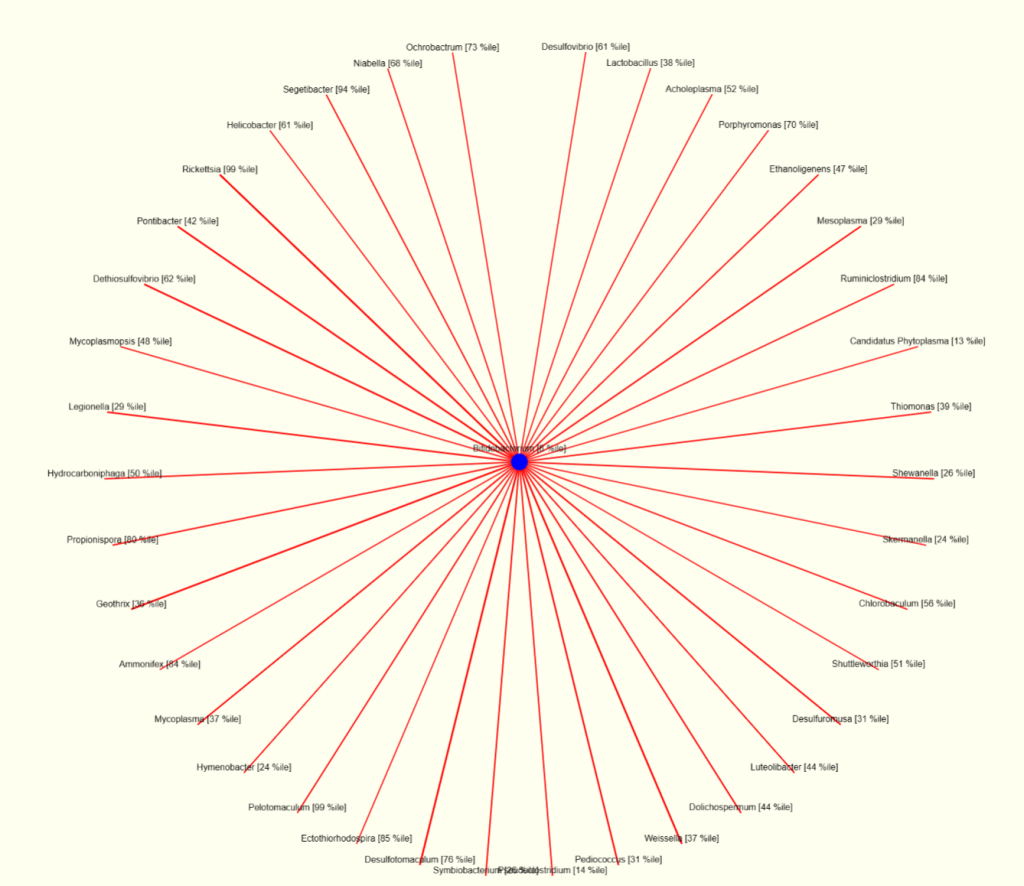
This appears to illustrate that being focused on just one or two bacteria may not be the best path. A holistic approach (looking at what changes all of the bacteria above may be a critical step of improving things).
Antibiotic path
Long COVID is very similar to ME/CFS. Looking at antibiotics suggestions, about 60% of them are commonly use bt ME/CFS specialists. These areL
- metronidazole (antibiotic)s[CFS]
- ciprofloxacin (antibiotic)s[CFS]
- clindamycin (antibiotic)s[CFS]
- minocycline (antibiotic)s[CFS]
- ampicillin (antibiotic)s[CFS]
My personal preference would be one round of metronidazole, a three week break then minocycline, another break and then one of the other ones. This is following Dr. Jadin’s Current Protocol for ME/CFS using antibiotics
Postscript – and Reminder
I am not a licensed medical professional and there are strict laws where I live about “appearing to practice medicine”. I am safe when it is “academic models” and I keep to the language of science, especially statistics. I am not safe when the explanations have possible overtones of advising a patient instead of presenting data to be evaluated by a medical professional before implementing.
I cannot tell people what they should take or not take. I can inform people items that have better odds of improving their microbiome as a results on numeric calculations. I am a trained experienced statistician with appropriate degrees and professional memberships. All suggestions should be reviewed by your medical professional before starting.
The answers above describe my logic and thinking and is not intended to give advice to this person or any one. Always review with your knowledgeable medical professional.







Recent Comments