Warning: This is for Advance Users and Microbiome Nerds
This year I have been updating KEGG data with an careful audit. The existing KEGG data was strain specific, and the numbers being generated were only for those strains. I have averaged out over the species that these strains belong to so I can compute KEGG data compensating for missing data. Most 16s labs are reasonable complete for species reports but report only a few strains.
The result will be more accurate estimates when I update all of the pages and data computed from the raw data upload.
I am a firm believer in openness — showing which study or source that I obtain the data from. This allows people to independently validate (or nit-pick applicability).
To demo how we can manually get this data…
Request information on an enzyme by entry key, i.e. https://www.genome.jp/dbget-bin/www_bget?ec:1.1.1.10 . From this fulcrum page we get:
- The compounds being produced (Product)
- The compounds being consumed (Substrate)
- Taxonomy — the bacteria having this enzymes
There are also links to a lot more information, while informative, not immediately relevant to Microbiome Prescription.
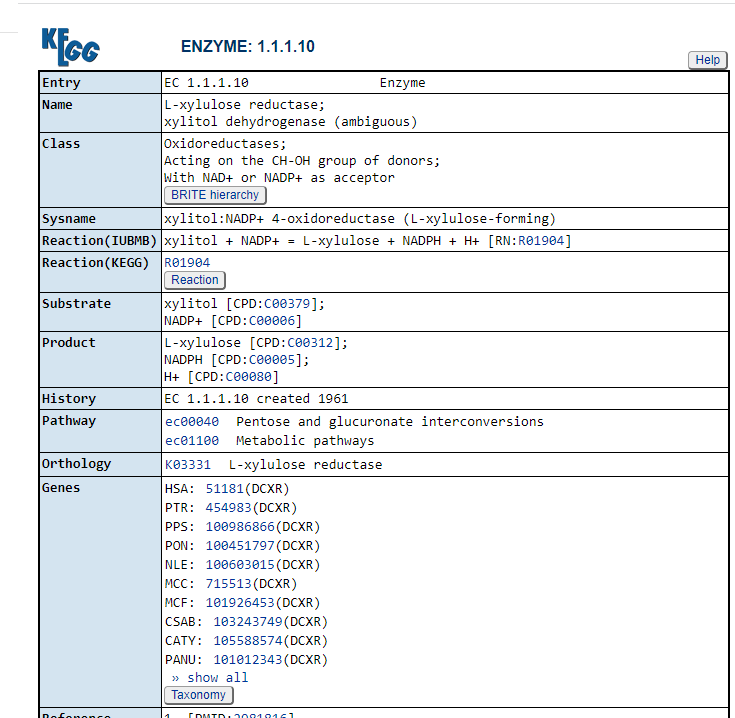
Clicking on the compounds (CPD: ….) takes you to a description of the compound
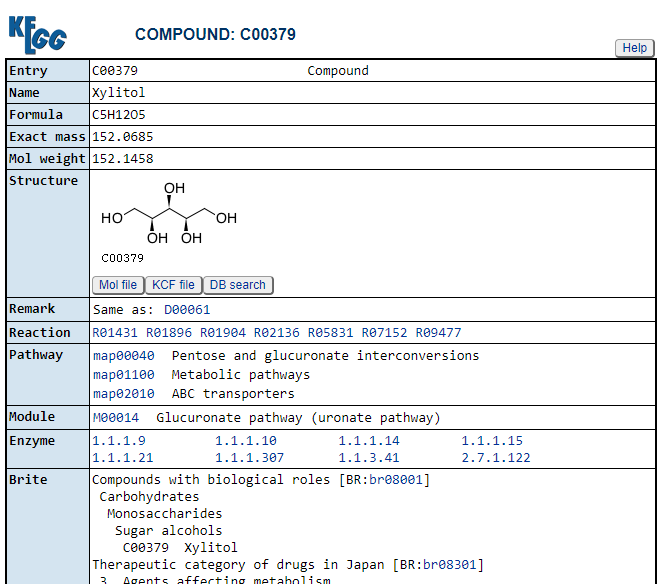
Then click on Taxonomy to get the bacteria (and other things it is found in). This will show you an expanding and collapsing tree. Look for Prokaryotes / Bacteria
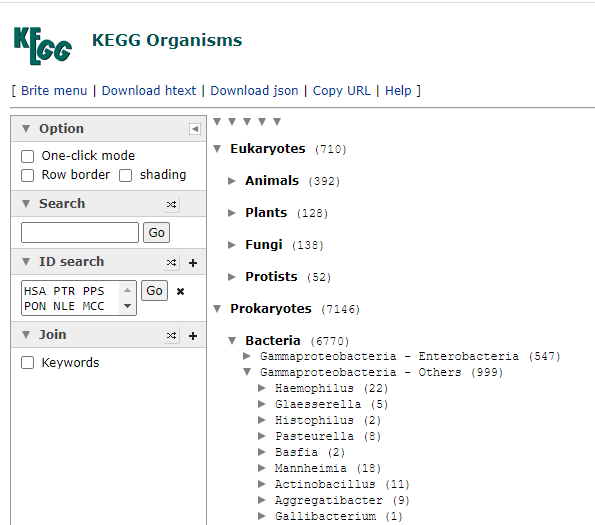
Expand until you find a strain of interest, all strains will be listed, it is the ones that are in blue, items in red do not have this enzyme. As you see below, this approach has some technical issues, one strain of Escherichia Coli has an enzyme and most of the others do not. To be technically complete, we would need the labs to identify every strain of Escherichia Coli as well as KEGG having all strains — that is unlikely for decades…
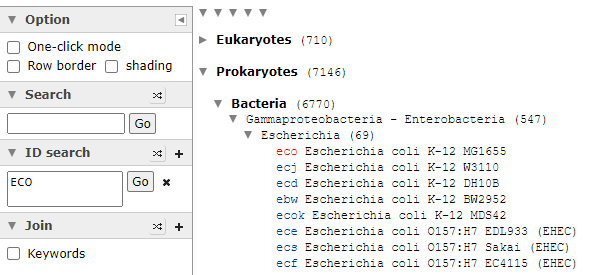
Clicking on it will show you the bacteria details as shown below with the all important NCBI Taxon number.
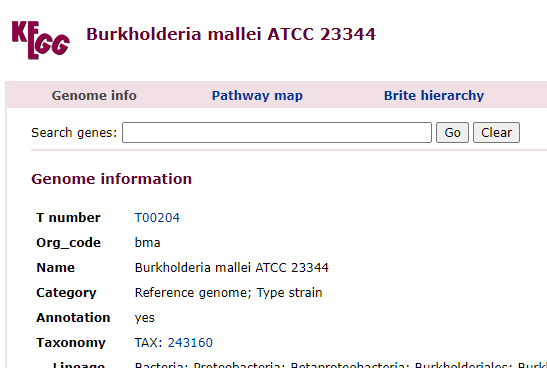
What is happening under the cover…
All of this data is extracted and re-aggerated into 3 main pages:
The images are below. Each page allows drill downs
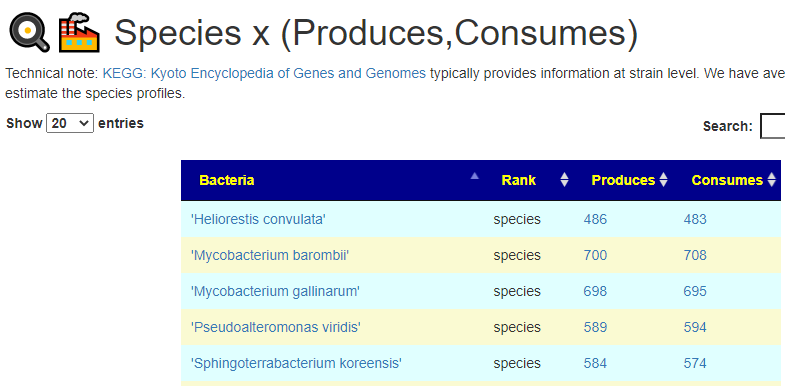
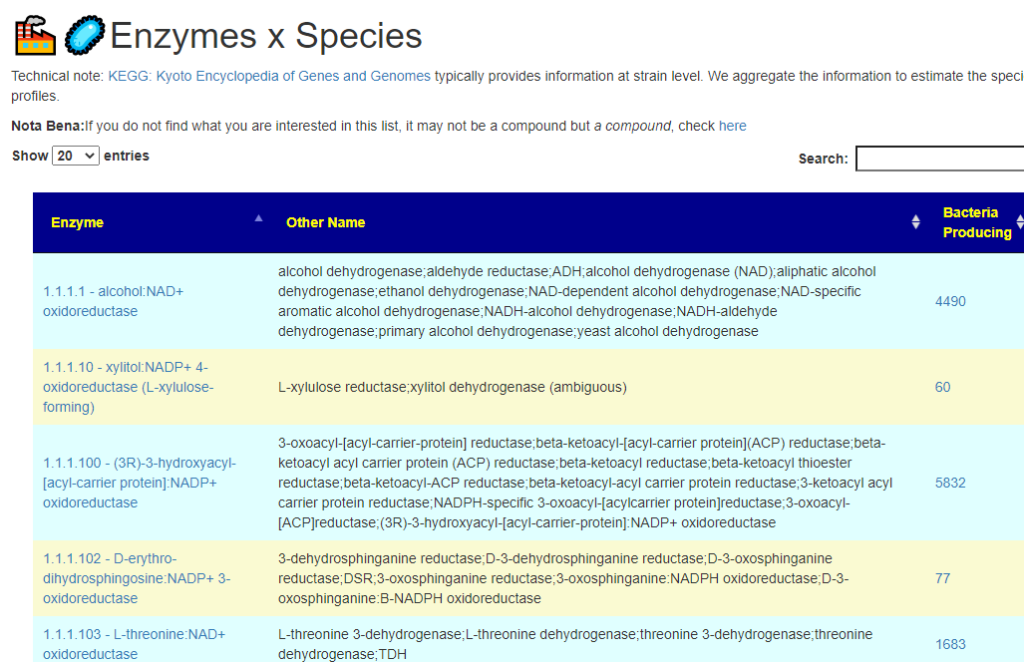
In the case of drill downs to bacteria, we also show a count of the number of enzymes in the bacteria that produces or consumes a compound, as illustrated below:

This is just step 1 – Showing the data
Step 2 is applying it to the samples and re-computing the totals and percentiles. Needless to say, many of these pages will be hidden on the menu to the casual user because it would result in information overload. An advance display level will be required. The above links should always work.
I still have some tuning to do, there are a few thing slightly off… but what is there is reasonably accurate.
Всех граждан в российской федерации а также белорусской республике, кто говорит за мир и также вперекор войны теперь за это можно объявить великими людьми