Hey do you think microbiota dysbiosis could cause circadian disturbance? Most articles go in an opposite direction and say its lifestyle causing circadian disturbance…But my disturbance is resistant to lifestyle… I just have primary circadian problem that might be even my worst symptom… Most resistant and almost lifelong.
Asked by a Reader
In keeping to “gold standard” of information instead of bloggers’ urban myths and ideologies, I head over to the National Library of Medicine studies.
- “gut microbial metabolites influence central and hepatic clock gene expression and sleep duration in the host and regulate body composition through circadian transcription factors”[2020]
- “Findings have suggested that gut microbiota play a major role in regulating brain functions through the gut-brain axis. A unique bidirectional communication between gut microbiota and maintenance of brain health could play a pivotal role in regulating incidences of neurodegenerative diseases. ” [2021]
Sleep, circadian rhythm, and gut microbiota [2020]
First, a more precise definition of circadian rhythms from the above study.
A fundamental part of eukaryotic life, circadian rhythms are endogenous, entrainable biological processes that oscillate in a 24-hour period in concert with the circadian environment of the earth. Circadian rhythms can be found at an intracellular level and have the ability to impact all aspects of metabolism (11). The mammalian circadian rhythm is orchestrated by a master clock, located in the suprachiasmatic nucleus (SCN) of the hypothalamus (12). The master clock follows the 24-hour light-dark cycle (the diurnal cycle) and coordinates the release of neurotransmitters such as serotonin and norepinephrine. Serotonin and norepinephrine are present at higher levels during wakefulness, while melatonin peaks during the night, regardless of the diurnal or nocturnal sleep cycles across species… The peripheral circadian clock is a system of organs within the 22 body which collect
environmental and internal signals in order to direct the expression of circadian clock genes
And then we read:
- “food intake can disassociate peripheral clock periodicity from the master clock; when this happens, greater immune system activation and metabolic dysfunction occur”
- “Dysbiosis and metabolic consequences resulting from circadian clock disruption may be due to increased permeability of the intestinal epithelial barrier “
- “gut microbial metabolites such as the short-chain fatty acids butyrate and acetate may influence clock gene expression“
- “Leone et al. found that a lack of gut microbiota, and consequently a deficit of microbial metabolites, resulted in markedly impaired central and hepatic circadian clock gene expression (40), suggesting the possibility that gut microbiota play a role in propagating circadian rhythm at the molecular level”
- “Serotonin deficiency elicits the loss of the circadian sleep-wake rhythm”
- “The microbes of the gastrointestinal tract exhibit circadian rhythm, and their composition oscillates in response to the daily feeding/fasting schedule.
The Role of Microbiome in Insomnia, Circadian Disturbance and Depression [2018]
The characteristics of the gastrointestinal microbiome and metabolism are related to the host’s sleep and circadian rhythm. Moreover, emotion and physiological stress can also affect the composition of the gut microorganisms. The gut microbiome and inflammation may be linked to sleep loss, circadian misalignment, affective disorders, and metabolic disease.
Circadian misalignment and the gut microbiome. A bidirectional relationship triggering inflammation and metabolic disorders”- a literature review [2020]
On the other hand, peripheral clocks are found in the nucleus of almost every single cell (eg, enterocyte, hepatocyte, myocyte, adipocyte), and they show circadian rhythms and oscillations that are dependent and independent of the circadian rhythms from the master clock. While the master clock responds mainly to light/dark cycle, peripheral clocks respond to other zeitgebers (eg, temperature, diet, timing, and content of food intake), which indirectly regulate the central clock …
However, Parabacteroides, Lachnospira, and Bulleida were specific to the human GI tract. Lachnospira was unique in that it was the dominant species that were affected by time and behavior (energy consumption early during the day) [114]. However, it is not fully understood why some species increase with clock time throughout the day. One of the theories is that some species are bile resistant, so they increase during the day as the food is ingested, and bile is secreted (eg, Oscillospira and )
A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host [2015]
“We found that up to 20% of all commensal species in mice and humans undergo diurnal fluctuations in their relative abundance, resulting in rhythmic changes of the entire bacterial community over the period of one day. For instance, the common mouse and human commensal genus Lactobacillus increases in relative abundance during the resting phase (the light phase in a mouse) and declines during the active phase.”
Bottom Line
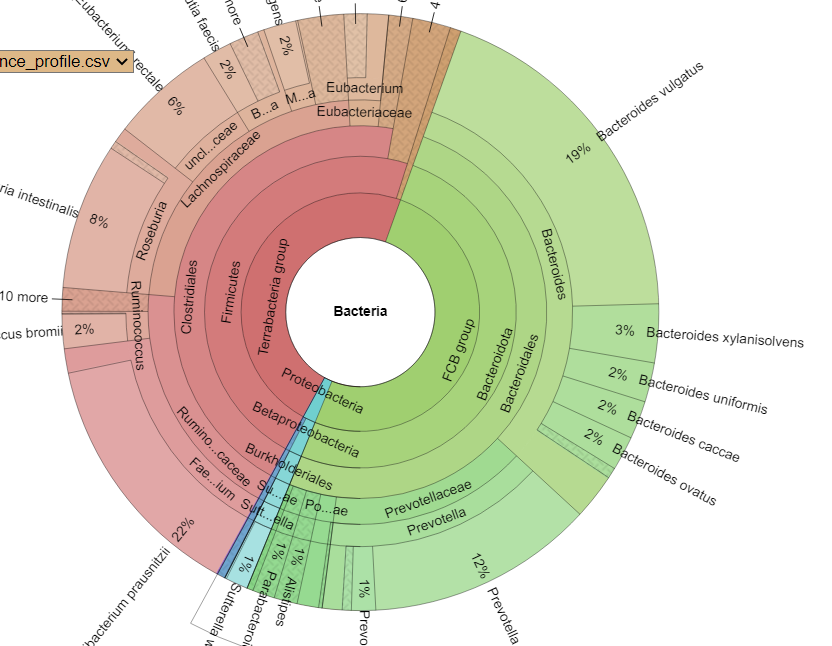
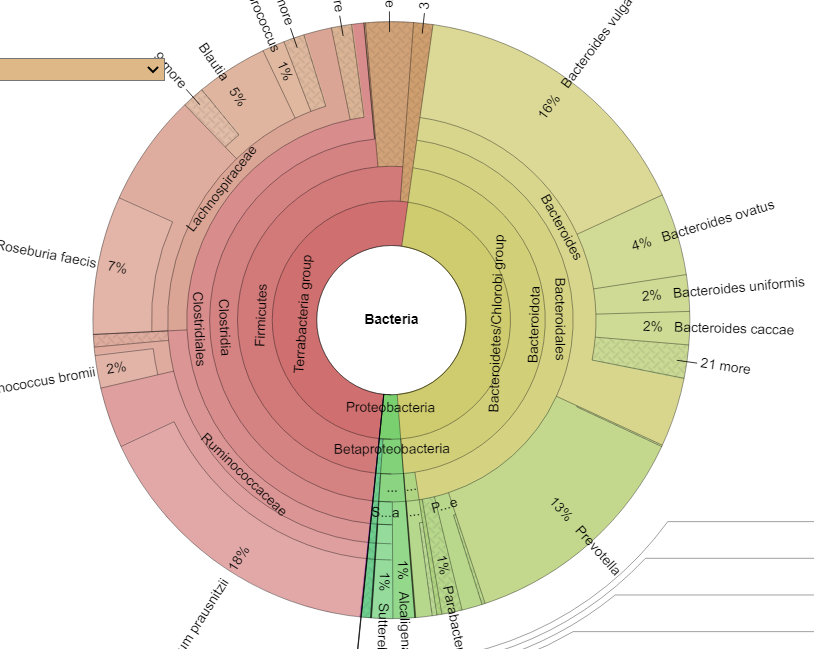
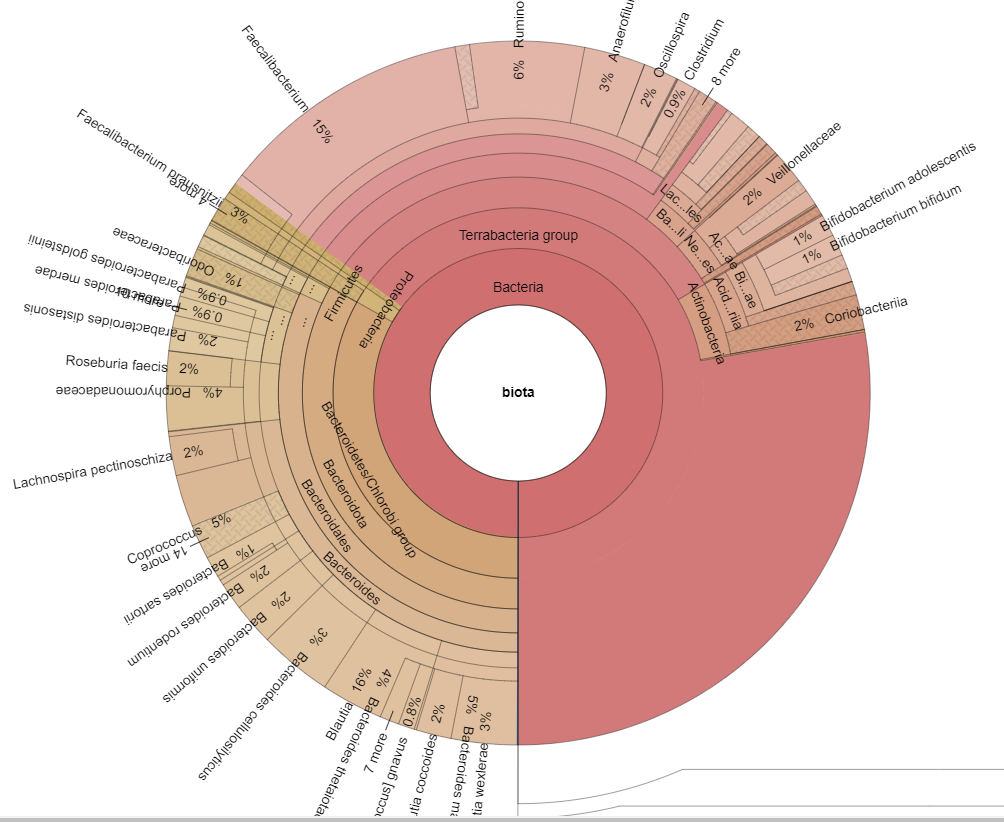
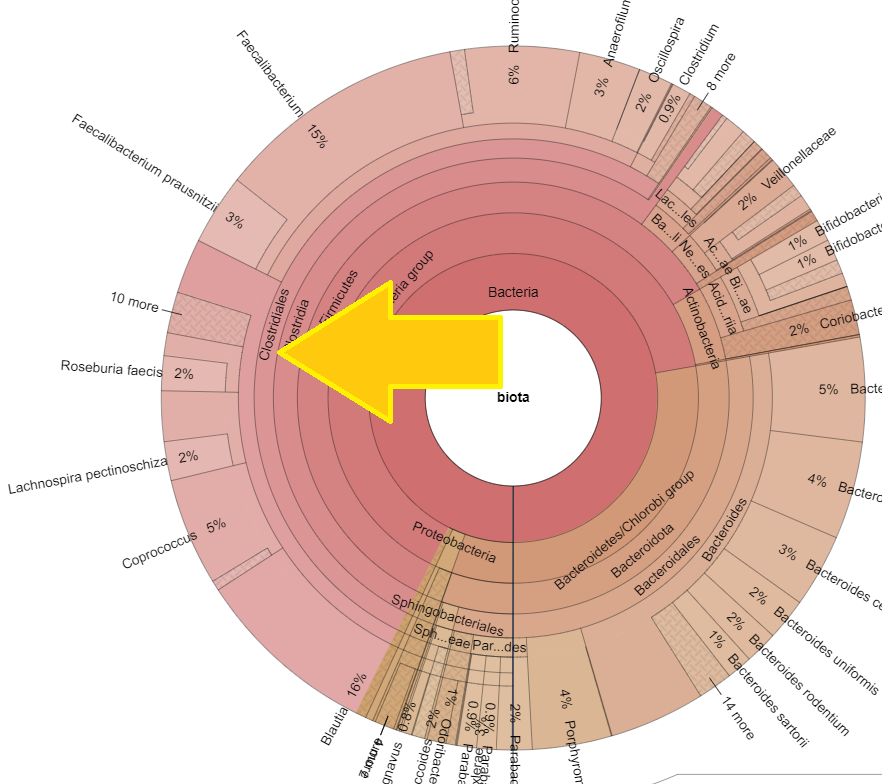
Time of day, time of year, eating time and diet impacts intra-day microbiome population and thus the metabolites being produced. Some of these metabolites have been shown to impact circadian cycle in recent studies. A few bacteria pulled from the studies cited above include:
- Fusobacterium
- Porphyromonas,
- Prevotella
- Bacteroides acidifaciens,
- Lactobacillus reuteri,
- Peptococcaceae
- Eggerthella,
- Anaerotruncus,
- Desulfovibrio,
- Roseburia,
- Ruminococcus
Time of year impacts (and may be a factor for Seasonal Affective Disorder – SAD)
- Helicobacter,
- Bacillus,
- Stenotrophomonas
- Proteobacteria,
- Lactobacillus
- Romboutsia
I was unable to find any 16s clinical studies on SAD
Advice for taking samples
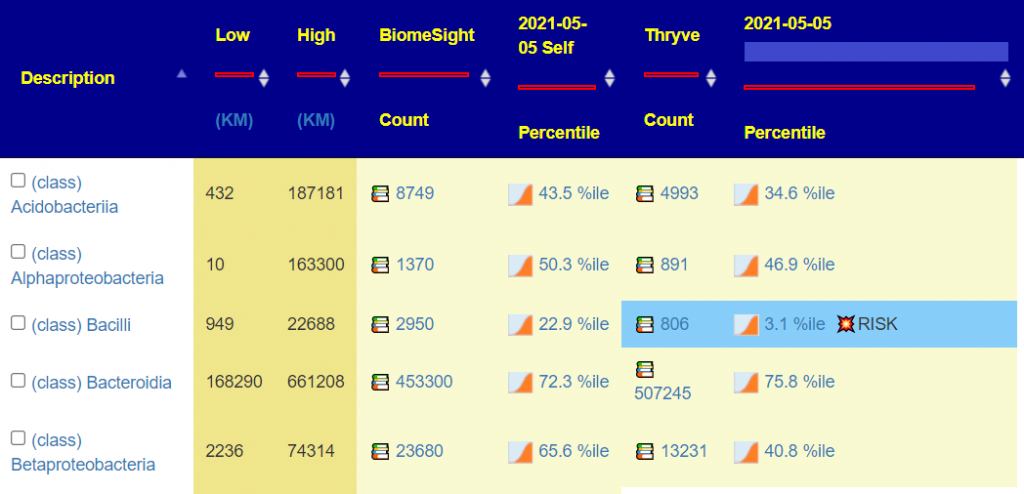
Record the day of the week, time of day, and if female, where you are in your cycle for stool samples. For best consistency (i.e. seeing what actually changed between samples) — make sure all follow up control for these factors as much as possible.







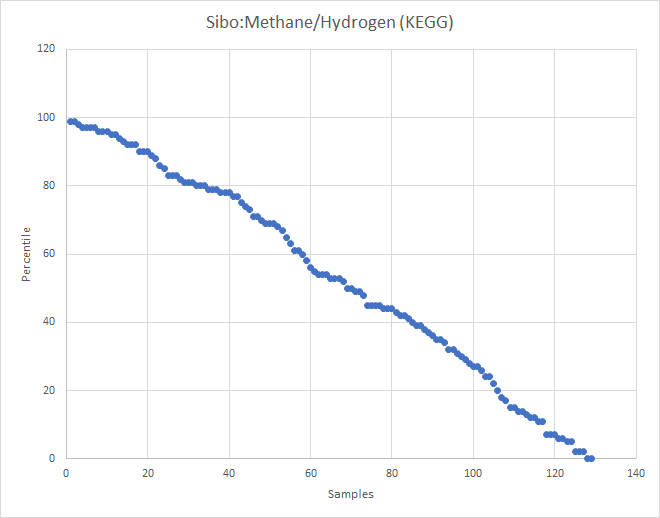
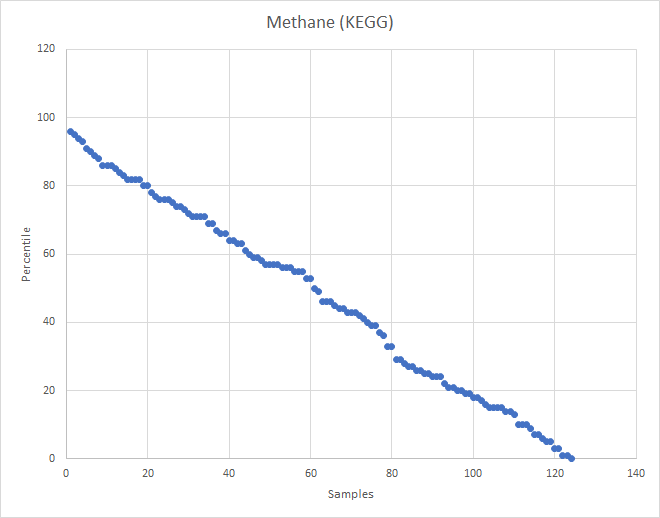
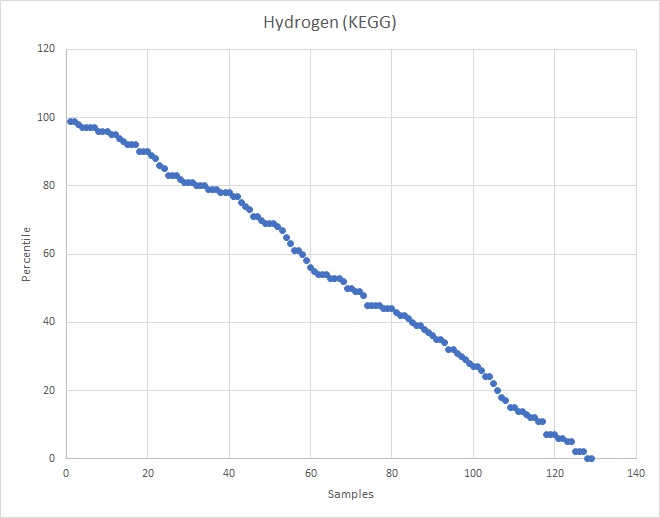
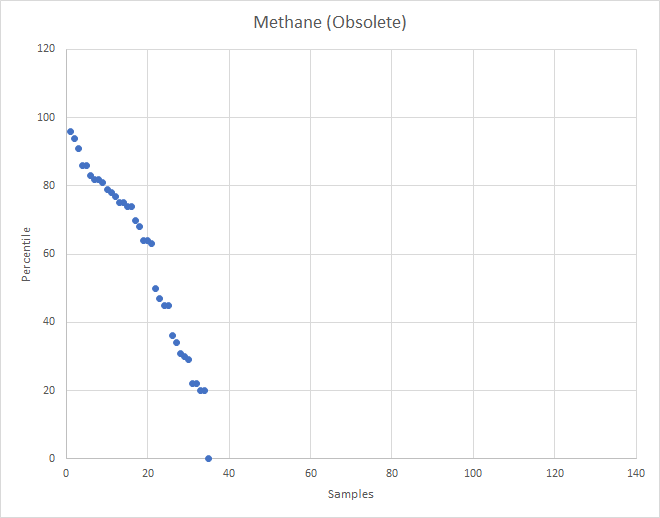
Recent Comments