I’ve recently added computations for Methane and Hydrogen using KEGG data to Microbiome Prescription. Checking the contributed symptoms, I had over 120 samples with SIBO indicated. So it was time to test the hypothesis.
Charts
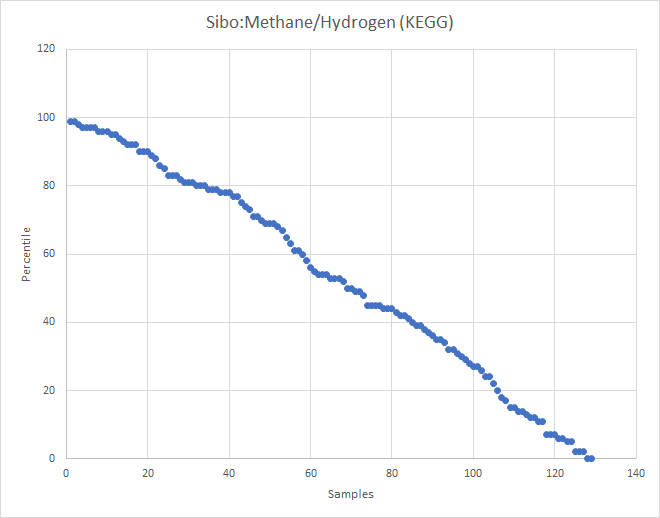
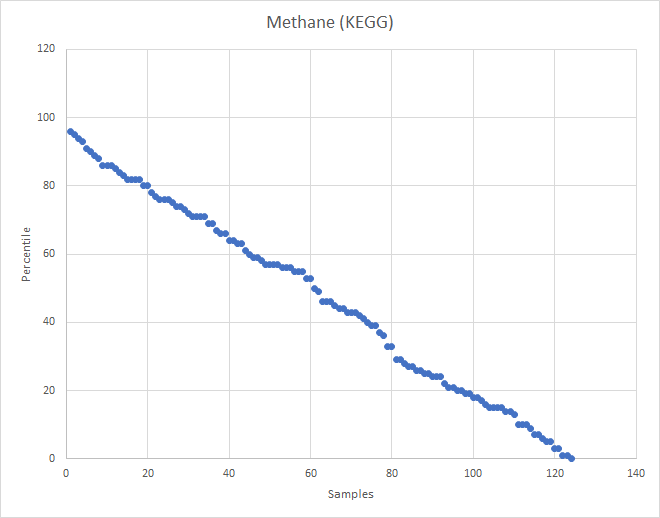
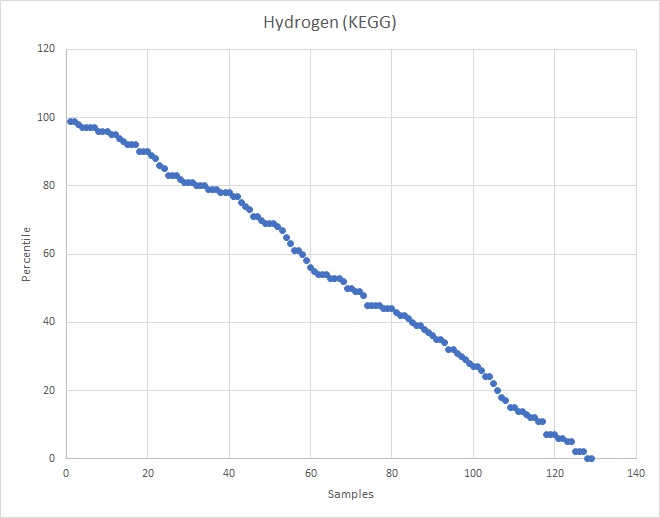
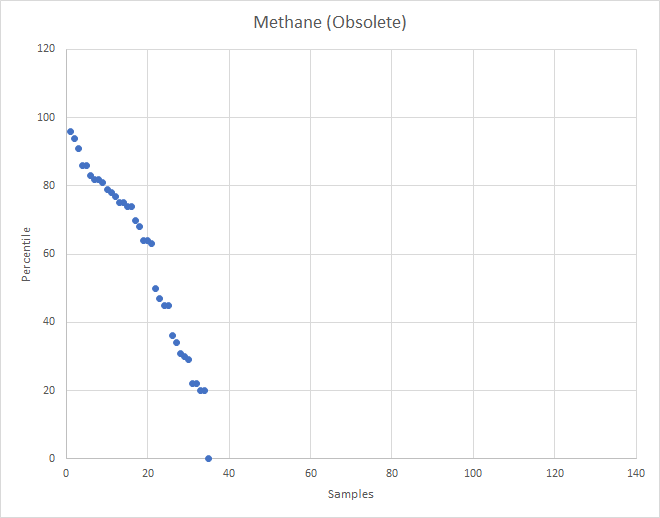
What about the old Methane?
The old computation was done on adhoc gathered associations from the literature. It also did not show any patterns.
Bottom Line
With the obvious path being unsuccessful, then time to examine where we did find associations.
| ProductName | Pattern |
| (4R,5S)-4,5,6-trihydroxy-2,3-dioxohexanoate | Between 33%ile to 66%ile |
| cob(II)yrinate a,c diamide | Between 66%ile to 100%ile |
| D-tagatose | Between 0%ile to 33%ile |
| undecaprenyl phosphate | Between 0%ile to 33%ile |
| EndProduct | Pattern |
| Vitamin B9 (Folic Acid/Folate) | Between 66%ile to 100%ile |
| Lactic acid | Between 0%ile to 33%ile |
| 2-Butanone | Between 66%ile to 100%ile |
| Hydrogen cyanide | Between 66%ile to 100%ile |
| Methyl thiocyanide | Between 66%ile to 100%ile |
| Propionate | Between 66%ile to 100%ile |
| Vitamin K | Between 66%ile to 100%ile |
| Vitamin B7 (biotin) | Between 66%ile to 100%ile |
| Sialic acid | Between 66%ile to 100%ile |
| Norepinephrine | Between 66%ile to 100%ile |
| EnzymeName | Pattern |
| succinyl-CoA:acetate CoA-transferase | Between 33%ile to 66%ile |
| phosphoenolpyruvate carboxykinase (GTP) | Between 33%ile to 66%ile |
| Definition | Pattern |
| Pentose phosphate pathway (Pentose phosphate cycle) | Between 75%ile to 100%ile |
| Pentose phosphate pathway, oxidative phase, glucose 6P => ribulose 5P | Between 75%ile to 100%ile |
| Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P | Between 75%ile to 100%ile |
| Serine biosynthesis, glycerate-3P => serine | Between 75%ile to 100%ile |
| Histidine degradation, histidine => N-formiminoglutamate => glutamate | Between 75%ile to 100%ile |
| Riboflavin biosynthesis, plants and bacteria, GTP => riboflavin/FMN/FAD | Between 75%ile to 100%ile |
| Tetrahydrofolate biosynthesis, GTP => THF | Between 75%ile to 100%ile |
| CAM (Crassulacean acid metabolism), light | Between 75%ile to 100%ile |
| Lysine biosynthesis, DAP dehydrogenase pathway, aspartate => lysine | Between 75%ile to 100%ile |
| tax_name | tax_rank | Pattern |
| Blautia hansenii | species | Between 33%ile to 66%ile |
| Filifactor alocis | species | Between 66%ile to 100%ile |
| Bacteroidia | class | Between 66%ile to 100%ile |
| Bacteroides gallinarum | species | Between 66%ile to 100%ile |
Last, we look at what studies reported
Looking at the “usual suspect” for SIBO, Methanobrevibacter smithii, a methane producer, we found only 23 samples with any (and all of the labs associated with these samples reports this bacteria so over 100 (80%) of the people reporting SIBO had none appearing)
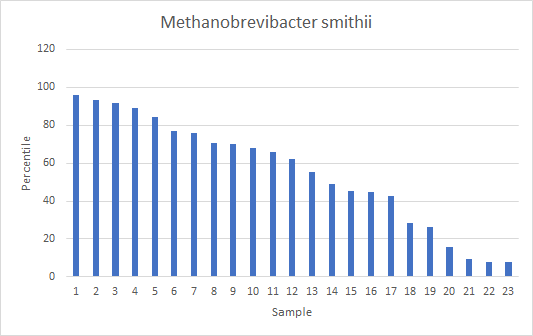
Tentatively conclusion, SIBO does not leave any clear tracks in a 16s Sample.
Recent Comments