Infrared Sauna and the Microbiome
Some 3 years ago, in this post I update my earlier post from 2013 and cited “, it is my hypothesis that this alters the microbiome — how, has still to be reported. “. There has been no objective impact on the human microbiome reported yet :-(.
Assuming that depression is partially microbiome caused (see this list of bacteria shifts seen with depression) then we have circumstantial evidence that they are changes (but do not know the details in humans). We do have information from one study on mice (see bottom of post)
- Feasibility and acceptability of a Whole-Body hyperthermia (WBH) protocol [2021] i.e. positive results for treatment for major depressive disorder
- Whole-Body Hyperthermia for the Treatment of Major Depressive Disorder: A Randomized Clinical Trial. [2016]
- The impact of whole-body hyperthermia interventions on mood and depression – are we ready for recommendations for clinical application? [2019]
- Changes in mood state following whole-body hyperthermia. [2992]
- Far-infrared Ray-mediated Antioxidant Potentials are Important for Attenuating Psychotoxic Disorders. [2019]
- ” FIR requires modulations of janus kinase 2 / signal transducer and activator of transcription 3 (JAK2/STAT3), nuclear factor E2- related factor 2 (Nrf-2), muscarinic M1 acetylcholine receptor (M1 mAChR), dopamine D1 receptor, protein kinase C δ gene, and glutathione peroxidase-1 gene for exerting the protective potentials in response to neuropsychotoxic conditions.”
- Far infrared radiation induces changes in gut microbiota and activates GPCRs in mice [2019]
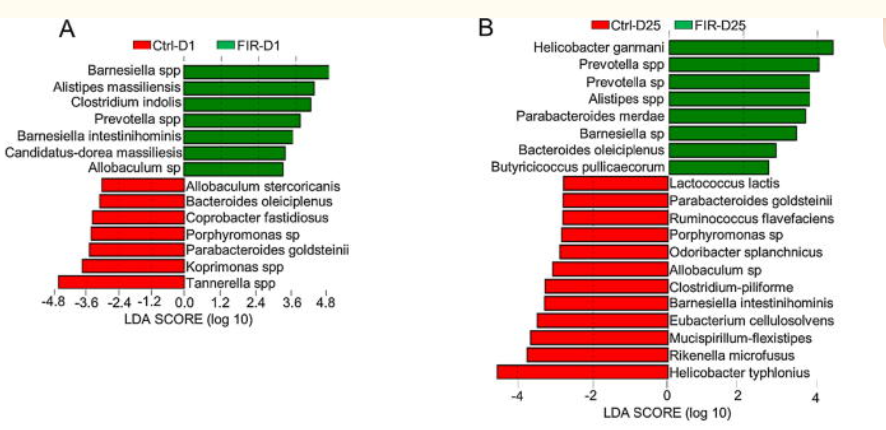
D1 – is Day 1, D25 is Day 25
Needless to say, the later information has been added to suggestions algorithms.
Picking Probiotics from OATS results
In a series of past posts, I walked thru the many pages of a OATS looking at each line:
- Organic Acids Tests (OATS) and Autism #1
- Organic Acids Tests (OATS) and Autism #2
- Organic Acid Tests (OATS) and Autism #3
- Organic Acid Tests (OATS) and Autism #4
A reader of that page presented me with a challenging question: “Which probiotic would reduce ….. ?” I checked the US National Library of Medicine studies — nothing. I am a lateral thinker (read Edward de Bono since I was a teenager) and it occurred to me that, theoretically, we can use data from KEGG: Kyoto Encyclopedia of Genes and Genomes because they have the gene sequence of many probiotics and thus their enzymes. Enzymes are mini-factories that consumes some metabolites and produces other metabolites. There are 5200+ different compounds reported on KEGG.
Since I have all of the data in a friendly (to me) datastore, it was just a matter of constructing a few complex queries and creating some web pages. The result was this page: Probiotics to Change KEGG Compounds
In the video below, I walk thru how we use OATS result and this page. Other test results can be used. OATS happened to be inspiration for this feature.
IBS + BioNTech COVID Vaccine -> ME/CFS?
A reader request a review of his results:
I am a 26 year old male living in Germany. I have a M. Sc. iIn 2018, I did a semester abroad in St. Petersburg, Russia, and during the exam period got IBS. I think stress and/ or vegan diet, which I only tried for a few months, played a role. Extremely low Vitamin D was found, but nothing else.
I developed a lot of food intolerances since then.
In April 2021 I got a Biontech vaccination, and in the following days, noticed that I was tired all the time. It did not get better. I was barely able to finish my Masters Thesis as it was almost finished, but could not start working. Long story short, I now have a lot of the common CFS symptoms, additionally my hair fell out and low testosterone was found.
From a reader
Some technical notes: It cannot be diagnosed as CFS because it has not lasted long enough. It can be viewed as post-immune reaction syndrome. Second, I too have concerns about post-immune reaction syndrome this year, to explain why:
- Three COVID 19 vaccinations
- Tetanus vaccination
- Pneumonia vaccination
- Two Singles vaccinations
After each, I saw my system “act up”, Some measurable — such as a jump in blood pressure that took a couple of weeks to calm down. More often, i was dragging for a couple of weeks. Night Sweats. skin inflammation, etc.
To Vac or Not to Vac — that is really not the question. It is equivalent to saying “I will not wear a seat belt in case the car plunges into a lake and traps me in the car” The risk of that happening is very low compare to the risk of not wearing a seat belt. There is no rationality that can supported by rational analysis.
Some studies showing that vaccination does alter the microbiome.
- Oral Vaccination against Lawsonia intracellularis Changes the Intestinal Microbiome in Weaned Piglets. [2021]
- Links between fecal microbiota and the response to vaccination against influenza A virus in pigs. [2021]
- Vaccination against the brown stomach worm, Teladorsagia circumcincta, followed by parasite challenge, induces inconsistent modifications in gut microbiota composition of lambs.[2021]
- Changes in the ceca microbiota of broilers vaccinated for coccidiosis or supplemented with salinomycin. [2021]
Human studies have likely not been done because they will be misused by “anti-vaccination” people. We can be confident that changes will happen. The nature of the change will depend on the prior state of the microbiome – an unstudied area. The change from each vaccine will likely be different.
I should mention that I have read several personal reports of major improvement of microbiome conditions as a result of vaccination. A percentage may go either way.
Where do we go from here?
We have 3 sets of microbiome changers – stress, IBS and vaccination. Food intolerance was something of interest — alas, I could not find anything on PubMed that identifies bacteria associated with it. Doing a quick scan of my Biome View, nothing really stood out.
- Holophagae class high
- Deinococci class high
- Deinococcus-Thermus phylum
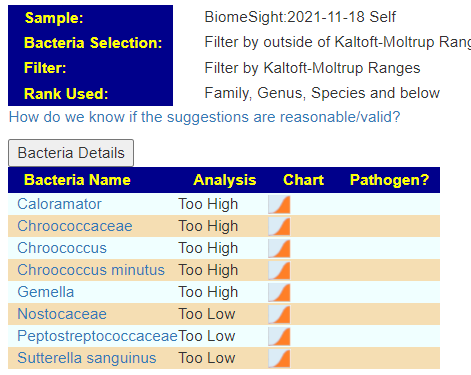
- Chroococcaceae family high
- Gemella family high
- Caloramator genus high (in terms of count — this was by far the highest of these)
Doing Due Diligence
- Quick ME/CFS choice returned not a single bacteria to be selected
- Advance Option IBS returned nothing with Kaltoft Moltrup,
- At 3%ile — two items were too low: Akkermansia muciniphila and Enterobacteriaceae
- At 6% — we added some highs: Lachnospira, Ruminococcaceae ( Ruminococcus)
Kaltoft-Moltrup Ranges
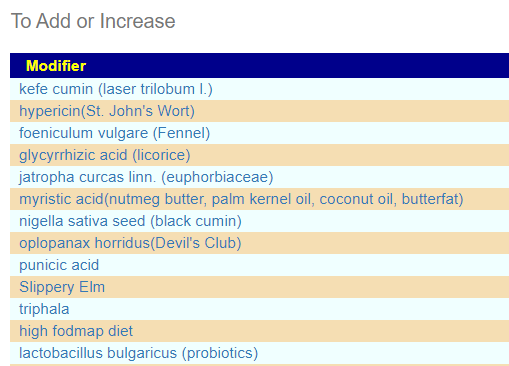
This produces a list FULL of unusual supplements… and a few familar. This is the first time that I have ever seen lactobacillus bulgaricus appear in the to add list. Some of my personal preferences (from experience with ME/CFS) are there: glycyrrhizic acid (licorice), Slippery Elm,triphala and quercetin,resveratrol
My general impression is that our list of usual suspects is really not there,
Time to Beat the Bushes
KEGG Generated Suggestions
The Weights were all below 20 — i.e. marginal, with Sun Wave Pharma/Bio Sun Instant, being the best of the short list
Similarly the supplement list was short at 10%ile and none at 5%ile
- beta-alanine
- D-Ribose
- L-Histidine
- Molybdenum
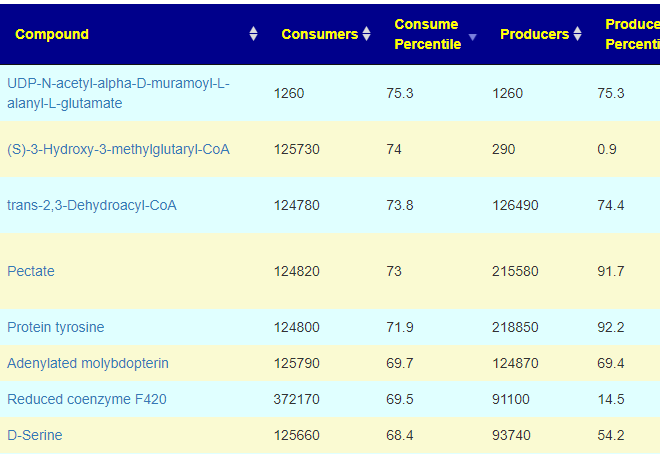
Compounds Produced and Consumed Page
Looking at the new Compounds Produced and Consumed, there were a number of items that had high ( > 99%ile) production – with 600 items listed, we would have expected just 6 (1%) not 41!!:
- ADP
- all-trans-Heptaprenyl diphosphate
- 5′-Deoxyadenosine
- 5-Phospho-alpha-D-ribose 1-diphosphate
- Acetaldehyde
- AMP
- CO2
- Diphosphate
- Fumarate
- Menaquinol
- NADH
- NADPH
- S-Adenosyl-L-homocysteine
- UDP
- Uracil
- Arsenite
- H+
- N-Acylsphingosine
- Xanthine
- Acetoacetyl-CoA
- Acyl-carrier protein
- D-Amino acid
- Protein lysine
- Protein-L-arginine
- (3S)-3,6-Diaminohexanoate
- 5-Methylthio-D-ribose
- Acetyl phosphate
- Alcohol
- D-Xylulose 5-phosphate
- Glycyl-tRNA(Gly)
- N-Formimino-L-glutamate
- Orthophosphate
- (S)-4,5-Dihydroxypentane-2,3-dione
- L-Sorbose
- N-Acetyl-beta-D-mannosaminyl-1,4-N-acetyl-D-glucosaminyldiphosphoundecaprenol
- 6-Deoxy-L-galactose
- Protoporphyrin
- tRNA
- D-Mannose 1-phosphate
- Spermidine
- Glycolate
In terms of consumers, the highest percentile was 75.3%ile, In fact most of the consumed items appear ‘balanced’
Proposed Model
Because events were recent, we have high volatility in the microbiome’s bacteria. I saw similar at the start of my relapse… the various clans of bacteria are fighting each other with short term victors of one group and then a reversal.
The apparent issue is massive overproduction of compounds!!! How to address that may mean that I need to add more code to identify the key bacteria responsible and thus the page changed as shown below. This is a logical but experimental novel approach.
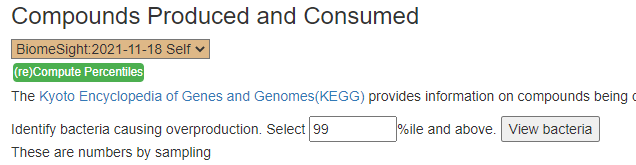
The result is shown below:
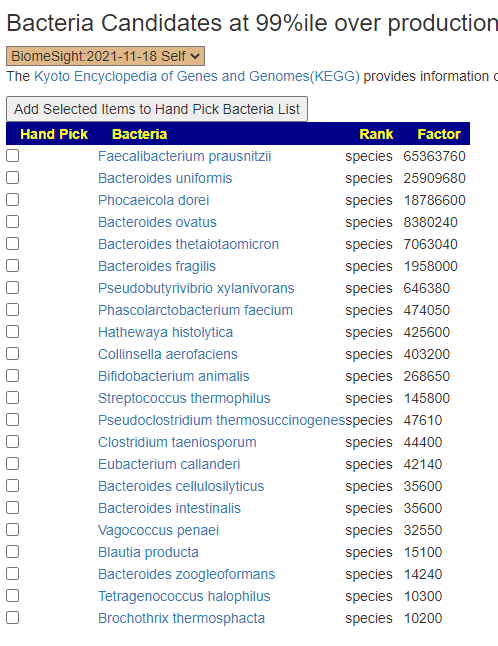
The result is show below. None were very high by themselves, Five were around 75%ile
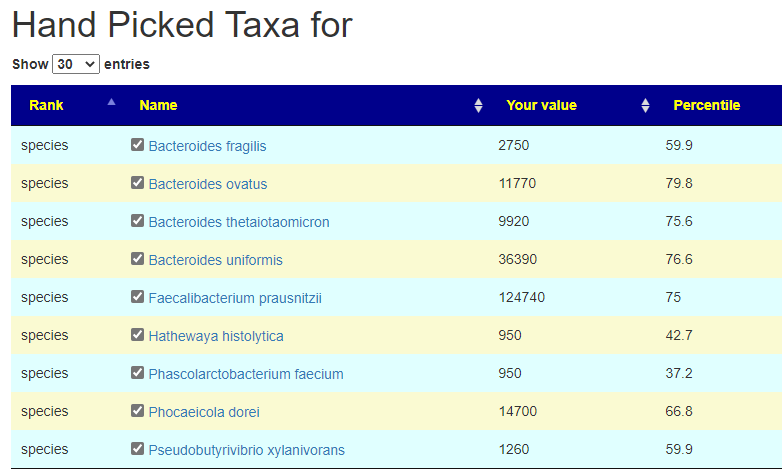
From this hand pick selection, we proceed to get suggestions. The results are shown below
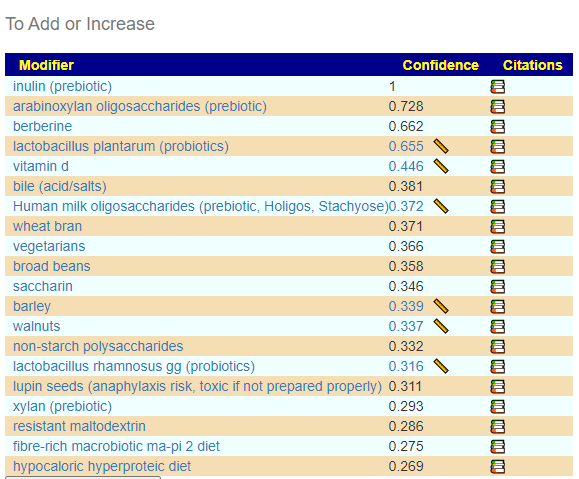
and the to-avoid
This looks similar to what I often see with ME/CFS people. That is, breakfast porridge made from barley with inulin and wheat bran with walnuts (which is my own regular breakfast!) with yogurt containing lactobacillus plantarum. There are some interesting studies in this area:
- Ingesting Yogurt Containing Lactobacillus plantarum OLL2712 Reduces Abdominal Fat Accumulation and Chronic Inflammation in Overweight Adults in a Randomized Placebo-Controlled Trial
- GABA enhancement by simple carbohydrates in yoghurt fermented using novel, self-cloned Lactobacillus plantarum Taj-Apis362 and metabolomics profiling
- Influence of Lactobacillus plantarum on yogurt fermentation properties and subsequent changes during postfermentation storage
- Probiotic yogurt containing heat-treated Lactobacillus plantarum enhances immune function in the elderly
Additional Lab Results from User
After getting to this point, the reader reported some recent lab results. Some are of interest:
- Vitamin D was just 28% into the normal range, so the vitamin D suggestion above is reasonable
- His coagulation factor II (G20210A/G) was at the high end of normal — I have the same coagulation issue and found that turmeric with black pepper or piracetam helps greatly — especially with brain fog and slowness.
- Protein S was low (barely in normal range), See Protein S deficiency
Issues causing hypercoagulation (thick blood) was shown to be common with ME/CFS by David Berg back in 1999, for articles and townhall transcripts (hosted by me!) see this page. This appears of part of this person’s causality.
Neither G20210A nor Protein S are likely to be deemed clinically significant, thus my personal preference (regular heparin taken sublingual, held for 1 minute and then spitted out) is unlikely to be prescribed.
For ways of addressing these, see my CFS Remission blog.
The COVID Vaccine Overtones
Prior to my getting my first COVID vaccine, I had concerns about it triggering coagulation – an ongoing ME/CFS risk. The reason was simple, vaccines triggers an immune reaction — milder than having COVID — but still an immune reaction. COVID was at that time, well known to produce coagulation issues (Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia April 2020) so they was a risk. The severity would likely be far less than that of COVID, but still enough to push someone with borderline coagulation issue across into ME/CFS. This appears to be correct as shown by some studies, a few are:
- First diagnosis of thrombotic thrombocytopenic purpura after SARS-CoV-2 vaccine – case report, Dec 2021
- Acquired thrombotic thrombocytopenic purpura: A rare disease associated with BNT162b2 vaccine Sep 2021
- Thrombotic thrombocytopenic purpura: a new menace after COVID bnt162b2 vaccine Jul, 2021
- Systematic Review of Antiphospholipid Antibodies in COVID-19 Patients: Culprits or Bystanders? [2021]
- Antiphospholipid antibodies in patients with COVID-19: A relevant observation? [2020]
This also is suspected with Long COVID,
- Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin Aug 2021
- Late-onset hematological complications post COVID-19: An emerging medical problem for the hematologist, Jan 2022
- and in The Guardian’s edition of today, “Could microclots help explain the mystery of long Covid?” Jan 5, 2022
Bottom Line
The microbiome is just a part of a health analysis, a significant part but far from being complete. We have a model for the tiredness. IMHO, non-prescription anti-coagulation treatments may eliminate it over a few months.
We are going to do two approaches that are connected.
- Coagulation — which appears to be caused by the bacteria of concern (a topic that I cover on CFS Remission)
- Bacteria — the ones that appear to be causing over production of many compounds
Reducing some of the bacteria cited above will likely also help, since many are known to cause coagulation:
- “These results indicated that Bacteroides sp. and F. mortiferum can accelerate blood coagulation in vivo ” [1973]
- “Bacteroides fragilis, Bacteroides vulgatus, and Fusobacterium mortiferum …. demonstrate that LPS of selected gram-negative anaerobes activate HF and thereby initiate the intrinsic pathway of coagulation.” [1984]
- Interaction of Bacteroides fragilis and Bacteroides thetaiotaomicron with the kallikrein-kinin system [2011]
- “Bacteroides fragilis and Bacteroides thetaiotaomicron, were found to bind HK and fibrinogen, the major clotting protein, “
- Unhappy Triad: Infection with Leptospira spp. Escherichia coli and Bacteroides uniformis Associated with an Unusual Manifestation of Portal Vein Thrombosis [2021]
Some questions from reader:
- “The only probiotic I will add is lactobacillus plantarum. (Or also lactobacillus rhamnosus gg?) I will have to rotate that. Here is the question: I would also like to address IBS. I read on cfsremission that some probiotics, like Prescript Assist, could lead to IBS remission. Prescript Assist was also 2nd place on the KEGG recommendations, although you said the weights were marginal. Would it be not unreasonable to try Prescript Assist at some point, to address IBS?“
- Yes, I would suggest two-four weeks on a probiotic and then rotate to the other
- “Does this plan sound reasonable?:
- Continue to take: D-Ribose, Vitamin D, Coenzyme Q10, Multi-Minerals, Magnesiummalate, L-Carnitine
- Discontinue: VSL3, Citrullinmalate, Vitamin C, Vitamin B12
- Add: As many of the recommendations as possible from the Kaltoft-Moltrup suggestions, the KEGG recommended supplements, and the novel approach. Add anticoagulants.
- After 2 months test again.
- Yes, if sound very reasonable – track objective measurements as much as possible
- New results IgA 1 is at 4500 with normal range 500-2000. This may be related to the vaccination but IgA is associated with a lot of things.
- Abrupt worsening of occult IgA nephropathy after the first dose of SARS-CoV-2 vaccination [2022]
- Need for symptom monitoring in IgA nephropathy patients post COVID-19 vaccination [2021]
- “Results in the naïve-vaccinated group, the mRNA-1273 vaccine induced significantly higher levels .. of IgA (2.1-fold, P < 0.001) as compared with the BNT162b2 vaccine.” [2021]
Dehydration caused mast cell and histamine issues
A reader forwarded this 2019 study Dehydration affects exercise-induced asthma and anaphylaxis [Oct 2019]. Soem quotes:
PubMed was searched from April to July of 2019 using predefined search terms “dehydration,” “exercise,” and “allergy responses.” Based on the reference search, more than one-hundred articles were identified
Also, numerous mast cells and eosinophils were recruited, therefore isotype switching to IgE antibodies occur, this hypersensitivity activates mast cell degranulation. After degranulation, proteases, leukotrienes, prostaglandins, and histamine lead to many kinds of allergy symptoms.
A few months ago, I wrote Hydration and the Microbiome which reviewed some literature.
The event that caused me to start looking at hydration, was a cheap home hydration device — a modern blue tooth scale, the one below. Currently $20.

As a result, because I was definitely dehydrated (but did not feel dehydrated — because that had been my new norm!) and proceeded to work on increasing hydration. At the start, I was 45.4% and have slowly worked it up to the current 47.2%. My goal is at least 57%, the middle of the normal range (ideally over 65%). My significant other has mast cell issues and was sitting well below 45% and is working on this change. One of the challenges is finding alternatives for items she takes that are known to cause dehydration — including antihistamines.
- Antihistamines, Blood pressure medicines, Chemotherapy, Diuretics, Laxatives. (U of Michigan)
So there is the appearance of a feedback loop: dehydration triggers mast cells, which result in the need for antihistamines, which then causes dehydration. The person is trapped!
A longer list from Oxford Medical:
- Fybogel, Lactulose, Macrogol, Senna, Bisacodyl, Docusate
- Antacids
- Ibuprofen, Naproxen, Diclofenac
- Loratadine, Cetirizine, Promethazine, Hydroxyzine
- Lisinopril, Ramipril, Losartan, Amlodipine, Felodipine, Bendroflumethiazide
In terms of the microbiome environment, we can see the impact from this study
Dehydration was accompanied by cell changes in solitary lymphoid nodules and Peyer’s patches. The proportion between lymphocytes, macrophages, and mast cells in lymphoid organs depended on the stage of dehydration. The inhibition of cell mitoses, disappearance of mature plasma cells and mast cells (per field of view), significant decrease in lymphocyte count, 4-5-fold increase in the number of destructive cells, and low density of cells and lymphatic network of the small intestine (per unit area) were observed on days 6 and 10 of dehydration. Severe morphological changes were also revealed in other layers of the small intestinal wall (mucosa, submucosa, lamina propria, etc.).
Effect of dehydration on morphogenesis of the lymphatic network and immune structures in the small intestine [2008]
Bottom Line
Hydration plays a very important role for health. Drinking 6 liters of water/day is the classic health advice. This may be a over simplification. This area is still being studied. “As the most effective sports nutrition supplement, sports beverage has different ingredients and formulas, and also has various effects. To provide clues for the development of sports beverage, this article reviews the types, components, effects, and mechanisms of sports beverage currently used in post-exercise fluid restoration.”[Research Progress on Application of Sports Beverage to Post-exercise Fluid Restoration, 2021] So some sports drink may help, some may have little effect.
An unexpected side effect, I did not change my diet habits but with the shift of hydration, I also lost 12 lbs (6 kilos) over 7 weeks.
Picking Probiotics based on Studies
Several pages has been added that should help people pick and understand probiotic impact better
Picking Probiotics for a Condition
Page: https://microbiomeprescription.com/Library/ProbioticSearch
This page will search over 3000 studies on the US National Library of Medicine for studies that:
- Used probiotic strains that are available as retail products
- Mentions the word you search for in the study
We then add the name of the product containing it, with a link to a site selling it. There may be other products that also include it.
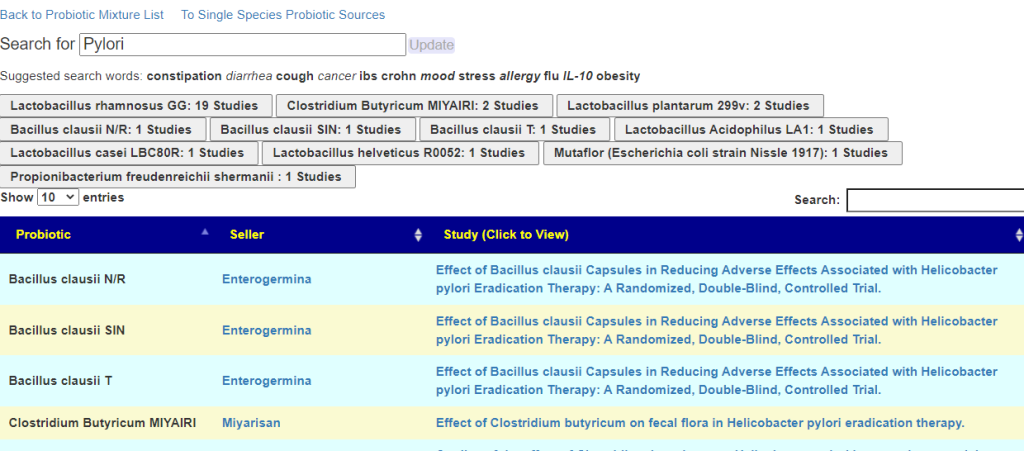
Why I react to one probiotic but not the other
This does a cascade, from the probiotic mixture we look at all of the species/strains in it, then we go over to the KEGG: Kyoto Encyclopedia of Genes and Genomes and lookup the enzymes being produced by these bacteria. From these enzymes, we look up the products being produced.
When you compare two probiotics mixtures, you may not that one produces a lot more of some products than the other. These are likely candidates for why you have a different response.
page: https://microbiomeprescription.com/library/ProbioticCompoundCompare
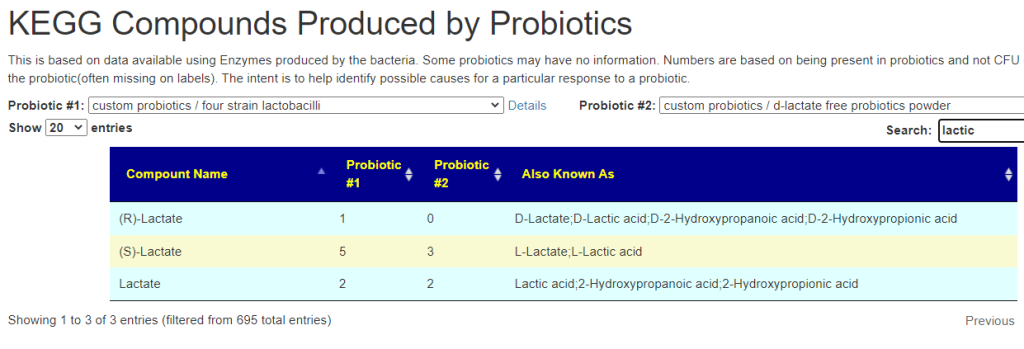
Walk Thru
Supplements for COVID
This is a special, off topic post resulting from some readers request. It is particularly important in the face of Omicron.
As always, I use gold standard sources:
Off-Label use of some Prescription Medicine
- Bacillus calmette-guerin [Tuberculosis] vaccination. [2021] decrease risk by 50%
- “Despite some controversies, numerous studies have reported a significant association between the use of ACE inhibitors and reduced risk of COVID-19″ [2021]
- “The results obtained from this study showed that Ramipril, Delapril and Lisinopril could bind with ACE2 receptor and [SARS-CoV-2/ACE2] complex better” [2021] – these are often prescribed for high blood pressure and appear to drop the levels of ACE2 significantly within a week.
Supplements
There is a mountain of speculation and few actual trials (Traditional medicines prescribed for prevention of COVID–19: Use with caution.). Often the criteria to suggest something is the belief that the substance will improve the immune function. This is actually a two edge sword — a stronger immune response can make someone sicker. Herbal Immune Booster-Induced Liver Injury in the COVID-19 Pandemic – A Case Series [Dec 2021]. Other suggestions are based on treating symptoms like coagulation with serrapeptase — suggested but not tested. Others on their impact on interleukins. virus titers, and interferon (IFN – for example from probiotics — again, suggested/speculated with no clinical trials.[See Probiotics Against Viruses; COVID-19 is a Paper Tiger: A Systematic Review [2021]
- “The findings from our study suggest that zinc supplementation in all three doses (10, 25, and 50 mg) may be an effective prophylaxis of symptomatic COVID-19 and may mitigate the severity of COVID-19 infection. “[2021]
- “This meta-analysis indicates a beneficial role of vitamin D supplementation on ICU admission, but not on mortality, of COVID-19 patients.” [2021]
- ” There is no data from interventional trials clearly indicating that vitamin-D supplementation may prevent against COVID-19.” [Dec 2021]
- “Evidence supporting the therapeutic use of HDICV(high-dose intravenous vitamin C) in COVID-19 patients is lacking.” [2021]
- Withania somnifera [Ashwagandha] showed the highest binding affinity and best fit … may reduce the glycosylation of SARS-CoV-2 via interacting with Asn343 and inhibit viral replication. [2021]
- Withania somnifera as a safer option to hydroxychloroquine in the chemoprophylaxis of COVID-19: Results of interim analysis [2021]
- “. Overall, the simulation run confirms the stability and rigidity of the interactions of Withanolide R and 2,3‐Dihydrowithaferin A from Withania somnifera to be the most potent phytochemical inhibitor for the main protease and the spike protein respectively. ” [2020]
- More studies
- “The study found a reduced risk of COVID-19 infection in participants receiving neem(Azadirachta Indica A. Juss) capsules” [2021]
Items with In Vitro Evidence that it may help
Remember that In Vitro often fails to reproduce with humans.
After Infection
In theory, some of these items may slow the infection by slowing the mechanisms of the infection.
- “Network Pharmacology Reveals That Resveratrol Can Alleviate COVID-19-Related Hyperinflammation” [2021[
- This search gives other interesting leads for substance that may help based on Network Pharmacology, many of them are herbs used in Chinese medicine. Some candidates are:
- Selenium [2021]
Bottom Line
If you find additional gold standard studies done on actual people, please forward them to me for inclusion on this page.
Rosacea, Circulation and mild CFS
Person Description
- 32 year old male living in Scandinavia
- Health problems started at 16 years(possible overtrainig).
- Symptoms:
- Every night my body collapses,
- my nose gets extremely red (rosacea),
- pulse rises and it feels strange in my body.
- Delayed if does not eat dinner.
- Feel very tired and can’t work out or even go short walks without getting tired (
- mild CFS – diagnosed by a healthcare practitioner at 25 years
- Brain fog.
- Frozen/poor circulation in the body.
- My body feels stressed,
- Always feel and hear my pulse in the ears/head.
- Problem with allergies/hypersensitivity to pollen, animals, strong fragrances, and against a lot of foods that I find difficult to identify.
- Starting to get a lot of grey hair (none of my parents got grey hair in such a young age).
- Diagnosed with SIBO 2 years ago.
- Diagnosed with Epstein-Barr virus 4 years ago.
- Every night my body collapses,
I have done a lot of different protocols but it none really help. Most beneficial for the red nose (rosacea) is eating a diet is consuming low meat, low fat, low starch and eat a lot of fruits, juices and smoothies.
Comment: My own experience dealing with a ME/CFS relapse was that they first test-change-retest cycles had little/slow progress (See my progress reports). He reported some positive change from the first cycle,
Microbiome Tests
First microbiome test in late August, 2021. Upload to Microbiome Prescription resulted in the following protocol:
- Week 1-2: Rifaximin (as I understand you are not a fan of Rifaximin, but it was one of my top modifiers and I am also diagnosed with SIBO and have huge issues with rosacea on the nose).
- Week 3-4: HMOWeek
- 5-6: Resveratrol, D-ribose (which I’m going to continue taking) and DAO.
- Week 7-8: Symbioflor 2.
From this changes, he reported a little more energy, but no real progress with my rosacea. Biomesight is processing his second test – will be ready in 1-3 weeks.
He is able to get antibiotics prescribed and He is also willing to take antibiotics.
Analysis
Rosacea
Rosacea is one of the conditions that I maintain in my US National Library of Medicine microbiome shifts list [rosacea]. The number of studies is few. In keeping with best practices (combining clinical studies with microbiome results) I checked Clinical Studies, I found Rosacea: Treatment targets based on new physiopathology data [Dec 7, 2021] lists “treatment targets and possible treatments:
- permanent vascular changes (medical and instrumental treatments);
- flushing (betablockers, botulinum toxin);
- innate immunity (antibiotics, nonspecific antioxidants and anti-inflammatory molecules);
- a neurovascular component (analgesics, antidepressants);
- Demodex (antiparasitic drugs);
- microbiome;
- skin barrier impairment (cosmetics and certain systemic drugs);
- sebaceous glands (isotretinoin, surgery);
- environmental factors (alcohol, coffee, UV exposure). “
So, approaching it via the microbiome is in agreement with latest clinical practice. We should be aware that “Frozen/poor circulation” and “vascular changes” hints at a possible co-factor.
- “there is a lack of well-designed and controlled studies evaluating the causal relationship between rosacea and dietary factors.” [2021]
- “Rosacea is triggered by hot and spicy food” [2021]
Reviewing literatures for Treatments and Clinical Studies
- Do not use metronidazole [2021]
- Possible doxycycline 40 mg/day – ongoing [2021] [2015] [2009]
- Minocycline 100 mg/day [2020]
- Gamma Linolenic Acid 320 mg/day [2020]
- “, mast cells (MCs) have emerged as key players in the pathogenesis of rosacea through the release of pro-inflammatory cytokines, chemokines, proteases, and antimicrobial peptides’ [2021] – hints at DAO and mast cell stabilizers
- silymarin/methylsulfonilmethane [2008]
- “Only 17% of those with rosacea were impaired by sunlight, whereas 26% improved. In the rosacea group, “[1989] – hints at Vitamin D
- “Serum vitamin D was lower in patients with rosacea” [2018]
- “The present study suggests that an increase in vitamin D levels may contribute to the development of rosacea. ApaI and TaqI polymorphisms, and heterozygous Cdx2, wildtype ApaI and mutant TaqI alleles were significantly associated with rosacea.” [2018]
- SHORT FORM: Positive or negative response to sunlight/vitamin D depends on DNA
Looking at Suggestions from 1st Analysis
For a starting point, I used Advanced Suggestions. NOTE: The database is constantly being updated so suggestions on the same sample may change over time.
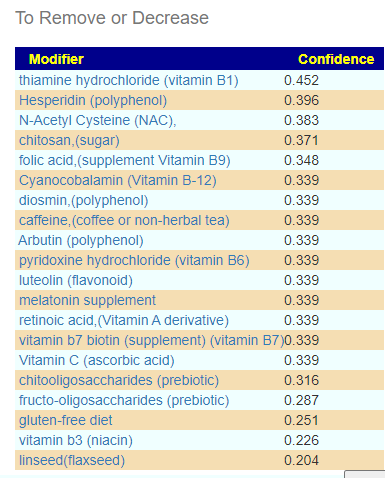
The results include some high weight items that was not included in his protocol. I checked if there was any results from clinical studies for Rosacea that used probiotics — I could not find any, thus the use of probiotics in a novel treatment approach. mutaflor escherichia coli nissle 1917 (probiotics) was included in take (so Symbioflor-2 is a valid substitution)
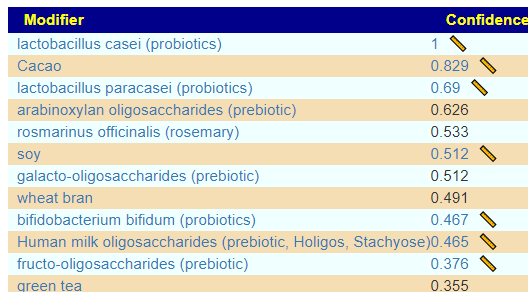
It is interesting that Lactobacillus casei and L. paracasei degrades histamines [Post 2021] which should improve allergies/hypersensitivity reported.
Please note that some probiotics are on the to-avoid list, including:
- lactobacillus reuteri (probiotics)
- bifidobacterium infantis,(probiotics)
- bacillus coagulans (probiotics)
- clostridium butyricum (probiotics)
- lactobacillus rhamnosus (probiotics)
- bifidobacterium longum bb536 (probiotics)
- enterococcus faecium (probiotic)
Dropping the Pub Med Filter for Rosacea, had some items that may impact circulation issues and mast cell issues positively:
Further Digging
The suspected cause being overtraining lead me to the shifts know to occur. The literature is below
| Citation |
|---|
| Intensive, prolonged exercise seemingly causes gut dysbiosis in female endurance runners. Journal of clinical biochemistry and nutrition (J Clin Biochem Nutr ) Vol: 68 Issue 3 Pages: 253-258 Pub: 2021 May Epub: 2020 Oct 31 Authors Morishima S , Aoi W , Kawamura A , Kawase T , Takagi T , Naito Y , Tsukahara T , Inoue R , Summary Html Article Publication |
| Rapid gut microbiome changes in a world-class ultramarathon runner. Physiological reports (Physiol Rep ) Vol: 7 Issue 24 Pages: e14313 Pub: 2019 Dec Epub: Authors Grosicki GJ , Durk RP , Bagley JR , Summary Html Article Publication |
| Physiological and Biochemical Effects of Intrinsically High and Low Exercise Capacities Through Multiomics Approaches. Frontiers in physiology (Front Physiol ) Vol: 10 Issue Pages: 1201 Pub: 2019 Epub: 2019 Sep 18 Authors Tung YT , Hsu YJ , Liao CC , Ho ST , Huang CC , Huang WC , Summary Html Article Publication |
| Improvement of non-invasive markers of NAFLD from an individualised, web-based exercise program. Alimentary pharmacology & therapeutics (Aliment Pharmacol Ther ) Vol: 50 Issue 8 Pages: 930-939 Pub: 2019 Oct Epub: 2019 Jul 25 Authors Huber Y , Pfirrmann D , Gebhardt I , Labenz C , Gehrke N , Straub BK , Ruckes C , Bantel H , Belda E , Clément K , Leeming DJ , Karsdal MA , Galle PR , Simon P , Schattenberg JM , Summary Publication Publication |
| Home-based exercise training influences gut bacterial levels in multiple sclerosis. Complementary therapies in clinical practice (Complement Ther Clin Pract ) Vol: 45 Issue Pages: 101463 Pub: 2021 Jul 30 Epub: 2021 Jul 30 Authors Mokhtarzade M , Molanouri Shamsi M , Abolhasani M , Bakhshi B , Sahraian MA , Quinn LS , Negaresh R , Summary Publication |
| Are nutrition and physical activity associated with gut microbiota? A pilot study on a sample of healthy young adults. Annali di igiene : medicina preventiva e di comunita (Ann Ig ) Vol: 32 Issue 5 Pages: 521-527 Pub: 2020 Sep-Oct Epub: Authors Valeriani F , Gallè F , Cattaruzza MS , Antinozzi M , Gianfranceschi G , Postiglione N , Romano Spica V , Liguori G , Summary Publication |
| Effect of an 8-week Exercise Training on Gut Microbiota in Physically Inactive Older Women. International journal of sports medicine (Int J Sports Med ) Vol: Issue Pages: Pub: 2020 Dec 15 Epub: 2020 Dec 15 Authors Zhong F , Wen X , Yang M , Lai HY , Momma H , Cheng L , Sun X , Nagatomi R , Huang C , Summary Publication Publication |
| The influence of exercise training volume alterations on the gut microbiome in highly-trained middle-distance runners. European journal of sport science (Eur J Sport Sci ) Vol: Issue Pages: 1-0 Pub: 2021 May 26 Epub: 2021 May 26 Authors Craven J , Cox AJ , Bellinger P , Desbrow B , Irwin C , Buchan J , McCartney D , Sabapathy S , Summary Publication |
Although it was a long time ago for the over-exercising, I decided to see if some of the changes may have the appearance of long term persistence. Running the known shifts against the two samples, I was actually surprised to see the earlier test had far more matches to the literature than the last test. Some items dropped off and only one item was added. Given that he had been implementing suggestions between the two samples, the improvement is nice to see.
| tax_name | tax_rank | Early Test Percentile | Last Test Percentile |
| Acidobacteria | phylum | 2.18 | |
| Actinobacteria | phylum | 7.85 | 3.91 |
| Akkermansia | genus | 0.71 | |
| Akkermansia muciniphila | species | 0.85 | |
| Burkholderiales | order | 26.90 | |
| Chroococcales | order | 3.24 | 26.00 |
| Clostridiaceae | family | 97.81 | 98.68 |
| Deferribacterales | order | 86.6 | |
| Eubacteriales Family XIII. Incertae Sedis | family | 14.75 | 6.56 |
| Hyphomicrobiales | order | 2.53 | |
| Moorella group | no rank | 2.54 | |
| Prevotella bivia | species | 75.47 | |
| Proteobacteria | phylum | 13.97 | 14.64 |
| Ruminococcaceae | family | 21.02 | 8.95 |
| Streptococcus australis | species | 87.87 | 83.60 |
| Streptococcus thermophilus | species | 83.16 | |
| Streptococcus vestibularis | species | 75.94 | |
| Sutterellaceae | family | 18.94 |
Continuing this logical exercise, “What would be suggested if I keep my scope to counteracting only the shifts that may be ascribed to over-exercising?” This assumes that the over-exercised shift created a stable dysfunctional microbiome that persisted. To do this, I used my Biome View and hand picked the above items. Many of the items are also on the list above.
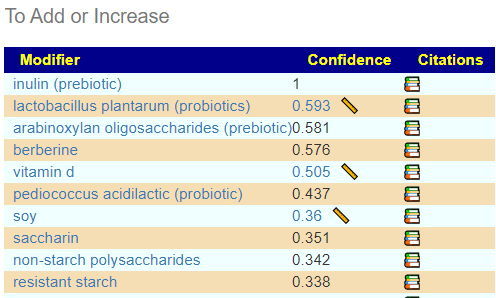
With one probiotic being well suggested (but likely difficult to get except by mail order). I included prescription items in the choices — none made it on the to add list, many appear on the to avoid list.
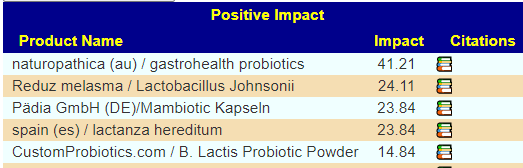
In terms of Flavonoids, Apples (I recall that at least 2 a day were needed from a study that I read recently) and Almonds (perhaps marzipan could work similarly… no studies, but given where he is, marzipan is definitely available)
Evaluating things tried
I recently added the ability to compare two samples in the light of taking some microbiome modifier (see this post for more details)
- Week 1-2: Rifaximin (as I understand you are not a fan of Rifaximin, but it was one of my top modifiers and I am also diagnosed with SIBO and have huge issues with rosacea on the nose). Results: AGREES 25, DIFFERS:23
- Week 3-4: HMO Week AGREES: 62 DIFFERS:58
- 5-6:
- Resveratrol, AGREES: 45 DIFFERS:40
- D-ribose (which I’m going to continue taking) AGREES: 6.2 DIFFERS:2.4
- DAO (Nothing in database)
- Week 7-8: Symbioflor 2. AGREES 0.5 DIFFERS:4.6
- I decided to checked Mutaflor (it’s siblings) AGREES 4.1 DIFFERS 2.4
The goal of this website has always been “Better suggestions than random suggestions”. In our review above, we found that AGREES > DIFFERS except for Symbioflor-2. The shifts seen more typical for Mutaflor (E.Coli Nissle 1917 probiotic). What I found most interesting was that the modifier that the reader has decided to continue, d-ribose, was also the one with the best ratio between Agrees/Differ. D-ribose has been documented to improve ME/CFS and FM (see this post for study links)
I was pleased to see almost every modifier had positive impact, but wish the amount was more.
Other Threads?
There is a tendency to try to fix everything at one time. My preference is to focus always on one to two items between tests. We have two threads — Rosacea and Over-exercising. Suggestions are similar and I would say run with it for 2-3 months and then retest.
As illustrated here (and in this prior review), we have some objectivity in evaluating the effectiveness of microbiome modifiers between samples — it should be helpful in improved picks over a series of samples.
Microbiome Fishing Expeditions
- If we review genus as an ethnic groups, we find over 3,365+ groups – each distinctive
- Species are the equivalent of small business, community groups – we have 10,567+
- Strains could be viewed as the individuals.
- Some strains (people) are criminals and some community leaders
- To understand what is happening in the cities, we need to get data / characteristics
- What foods are sent to the city
- What products are exported from the city
- What are the sources of energy for the city
- How may people come in or leave by ethnic groups
- What type of waste is produced by the city
- What types of crimes happen in the city
From this information, we may gain insight into how to administer and change the city.
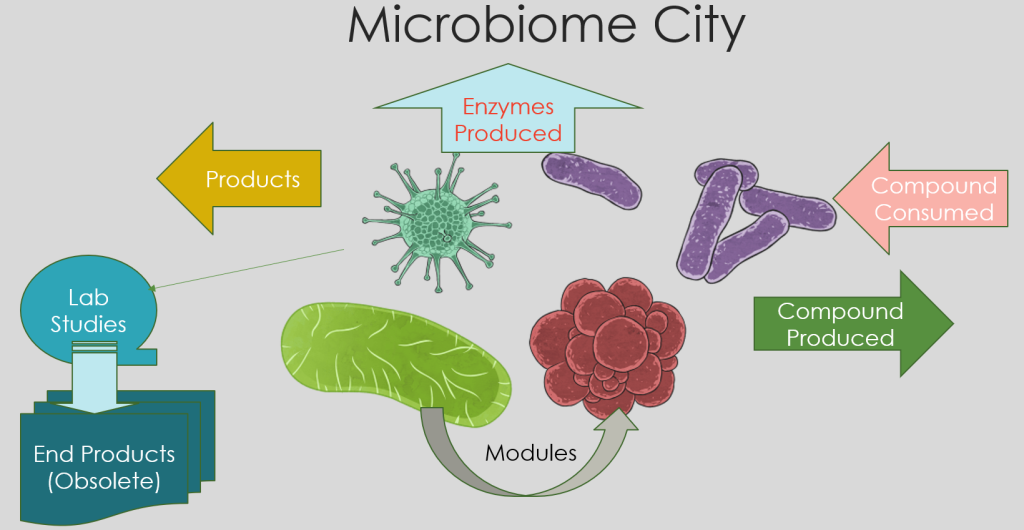
All of the above, except for End Products(Obsolete). This information is obtained from lab studies cultivating bacteria. Data is really hit and miss.
All of the other statistics is done using gene data from KEGG: Kyoto Encyclopedia of Genes and Genomes. Once the bacteria has been sequenced, all of this information can be computed. This works for both bacteria that can be cultured and those (most) that cannot be cultured in a lab.
Where do you find this data on Microbiome Prescription?
Seeing changes between Samples
When you have multiple samples, this line will appear. Multiple Samples Comparison is the button
These reports are the same as above – except you have numbers from different samples side by side
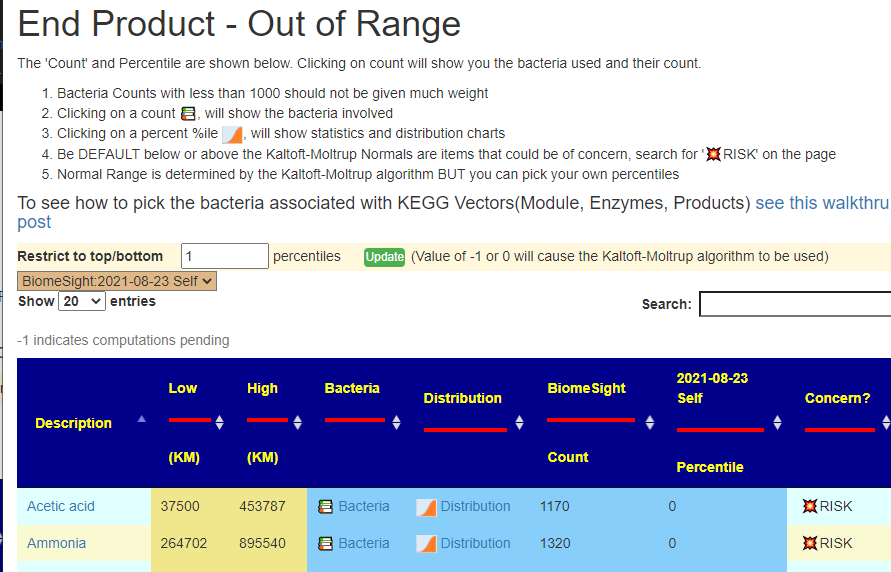
What is the difference between Outliers and Full Report?
Outliers ( AKA out of range) gives you the ability to filter by percentile OR use the Kaltoft-Møldrup Limits
The full range gives a much larger list (often hundreds) with filtering limited to the Search box on the page.
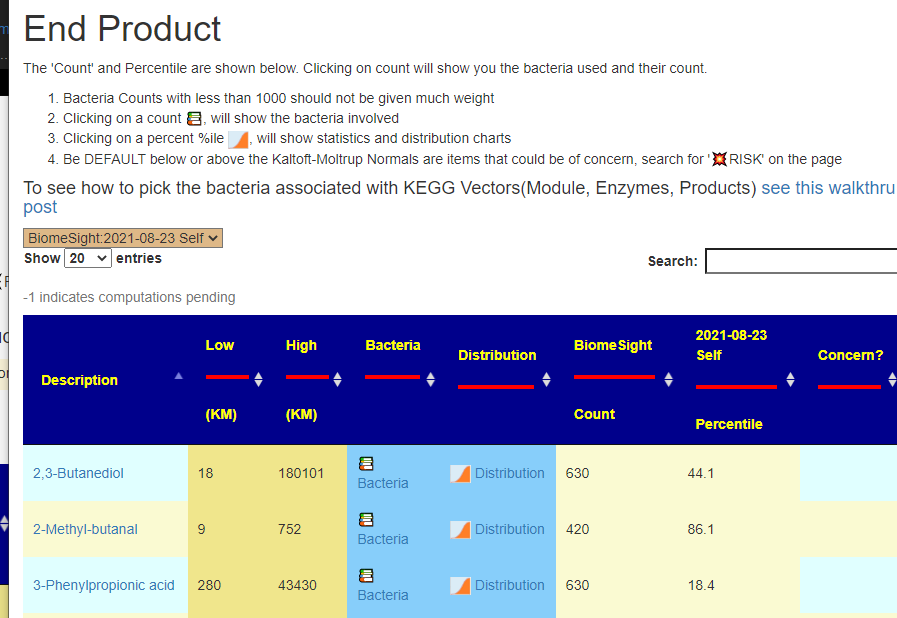
Above is for an individual sample — so you are given a list of your samples and the page will automatically update when you change samples.
For the multiple sample compare, you have already selected the samples and cannot change them from the page. You can go back to the selection page and pick different samples.
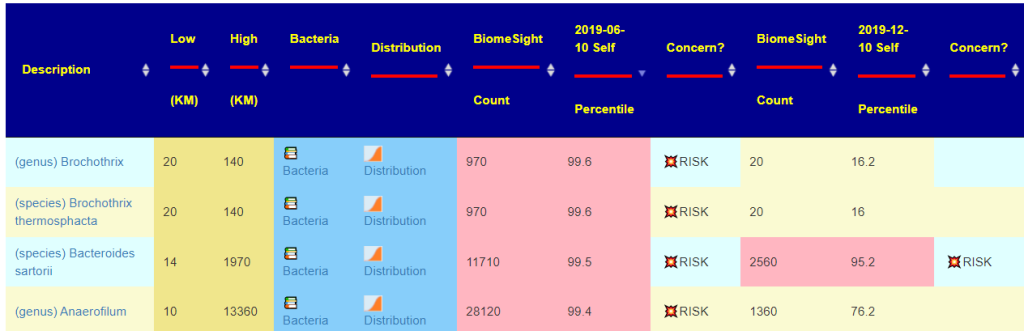
For multiple sample outliers — if one sample has an outlier, then the values of other samples will also be shown. The outliers are color coded:
VIDEOS COMING
A series of samples overtime from a CFS/ME person
The reader provide the following history (shortened)
Person’s Narrative
Sample BiomeSight:2021-02-24 In addition to my usual CFS symptoms I had a lot of trouble with gastritis. I was taking Slippery Elm, Marshmallow root, DGL, melatonin, magnesium, probiotic Bio.me Femme UT. I mostly avoided other supplements due to gastric issues. When the results came back I was only able to eat a few foods, so I couldn’t really follow the suggestions. I decided to wait until my stomach got better and retest.
Sample BiomeSight:2021-08-23 Taken after I’ve been on PPIs for 3 months. I haven’t been eating dairy for months which decreased B. Wadsworthia somewhat. I ate a lot of soy milk and soy yogurt. Mostly gluten-free, but eating lots of oatmeal. The combination of PPIs and yogurt made Streptococcus Thermophilus very high. The supplements at the time were melatonin, magnesium, zinc carnosine, d3 & k2, vit A, Omega 3, probiotic Bio.me Femme UT, occasional Slippery Elm to calm the stomach.
After I got results back I started taking:
– Miyarisan (Kegg AI Computed Probiotics),
– glycine (Kegg AI Computed Supplements),
– inulin, burdock root, L. Rhamnosus, L.Reuterii, almonds, oregano tea (suggestions from Targeted bacteria based on symptom Impaired memory & concentration).
I alternated modifiers so I wasn’t taking all of them all of the time.
I stopped:
– vit A and Slippery Elm (suggestions from Targeted bacteria based on symptom Impaired memory & concentration)
– yogurt (high S. Thermophilus)- oatmeal (it seemed to cause hypoglycemia. Replaced with tsampa – roasted barley flour)
Interestingly, Kegg AI Computed Supplements at level <15% reported Iron and 6 weeks later I was found to be anemic. That was an excellent prediction! I wish I would have gone and tested my iron status right away.
Sample BiomeSight:2021-11-28 Still on PPIs. Taken one month after I started taking prescription iron pills and lactulose to help with resulting constipation. Still eating soy milk daily, but no dairy, no yogurt. Taking more supplements at this point.
Analysis
Reading the history, my first question is to see if the predicted shifts from doing PPI (Proton Pump Inhibitors) is reflected between samples. Keep in mind that there are other microbiome compounders (i.e. other eating pattern changes). To this end, I’ve created a new page where you can pick a before, after and a modifier.

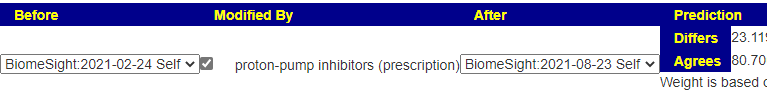

What is interesting to note is that the impact diminished overtime (80 -> 65). This agrees with my base model that rotation and re-testing is essential.
We can do combinations, for example PPI and melatonin between first and second sample. We lack studies on combinations and thus have to resort to this experiential approach to see how well predictions agree with actuals.

At the bottom of the pages, you get the details:
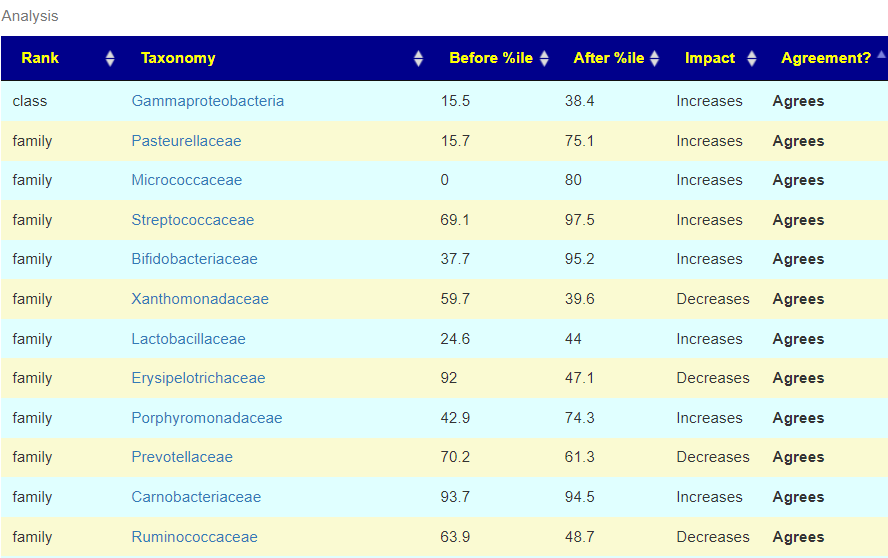
If a substance did not cause more improvements(Agrees versus Differs), then they are possible items to drop. Remember, the suggestions are theoretical prediction looking at many hundreds of bacteria. This allows you to objectively measure whether they worked well for you! They will not work for every one because of differences in DNA, diet, gender, age etc. I spot checked items like Slippery Elm (46.8 Agrees, 27 Differs), Miyarisan (KEGG Suggestion) (40.8 Agrees, 15 Differs) and was pleased with the results.
Looking at the next Step
Above, we (to my delight) verify that the suggestions are causing changes in the predicted direction for this person (based on their microbiome samples). Looking at predicting symptoms, we had 72% correct for the top 11 items ( So where do we go from here? Remember, the goal is to focus on the bacteria that likely contributed the most to the dysfunction (often high ones)
- From KEGG, the best probiotic is Sun Wave Pharma/Bio Sun Instant, if it is available. It has several species suggested in it.
- Looking at KEGG Products out of range — 7 were too low and 190 were too high. We are looking at an overproduction scenario.
I looked thru the predicted symptoms bacteria and there were a lot of secondary matches(shown with a # but very few direct matches. When we moved on to KM outliers, we had 25 items listed — 2 families and 5 genus with these items of considerable note — both for high percentile and high numbers:
- (family) Oscillospiraceae – 99.8%ile -18%
- (genus) Streptococcus – 98%ile – 4.2%
- (genus) Phocaeicola – 99%ile – 3.6%
So I did a hand picked with just these more extreme values. I tossed in everything we had. No prescription items showed up on the to take, but many (and a few b-vitamins) showed up on the to avoid list.
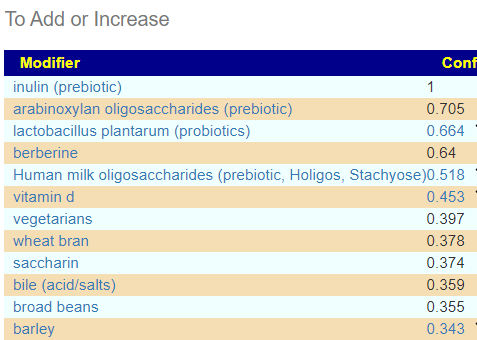
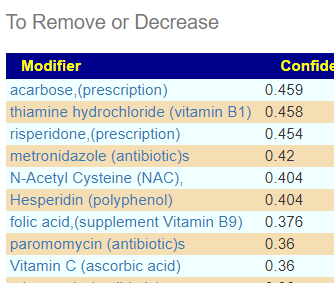
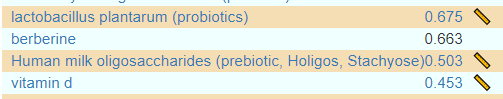
In terms of typical ME/CFS, we found only two matches, both two high: Bacteroides ovatus and Streptococcus. In terms of this small subset — only one item was above 0.29 — Human milk oligosaccharides (prebiotic, Holigos, Stachyose) with iron and B-12 being on the to take list.
We also note that the Unhealth Bacteria page, was full of Streptococcus
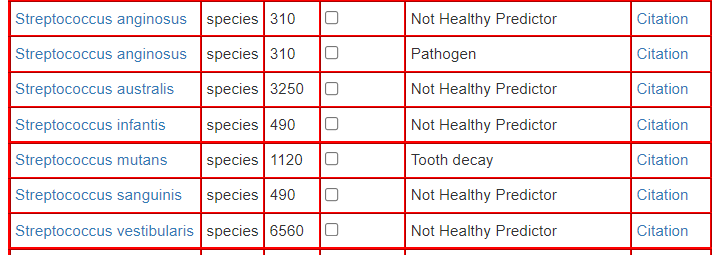
I tossed all of these into our hand-picked list and ask for new suggestions — the suggestions did not change expect for minor shifts of confidence.
Bottom Line
Two probiotics: lactobacillus plantarum (probiotics) and the mixture Sun Wave Pharma/Bio Sun Instant. I see the recommendation leads to the same type of breakfast that is regular for me: Barley/Oats with inulin and wheat bran. For all of the items, make sure you check the dosage links (where available)
Recent Comments