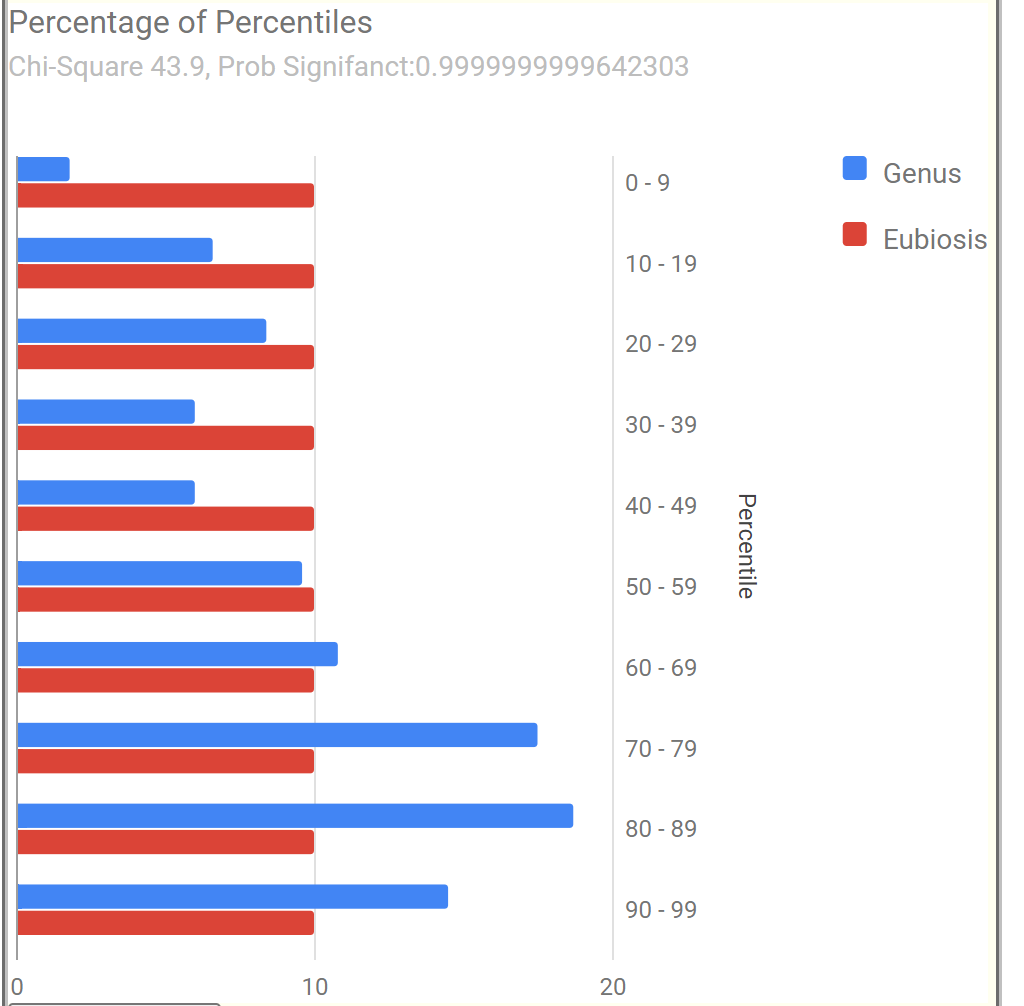
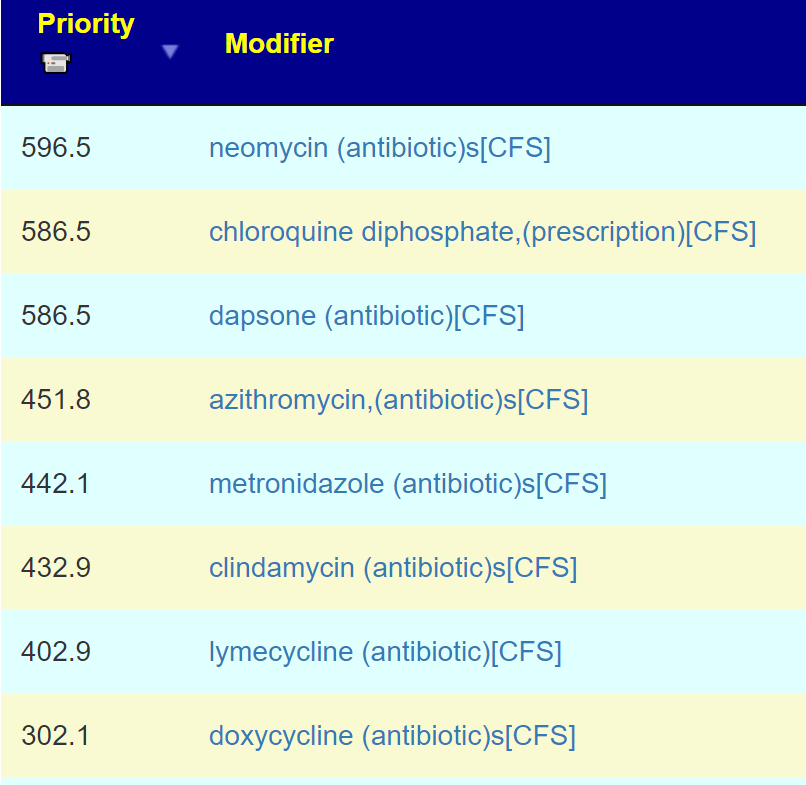
| Symptom Name | Factor |
| Neurological-Sleep: Sleep Apnea | 30.42 |
| Comorbid: Sugars cause sleep or cognitive issues | 29.39 |
| Comorbid: Panic Attacks | 29.30 |
| Comorbid: Carbohydrate intolerance | 28.89 |
| Official Diagnosis: Hypercholesterolemia (High Cholesterol) | 28.51 |
| DePaul University Fatigue Questionnaire : Fever & Chills | 28.14 |
| Neurological: Disorientation | 27.99 |
| Comorbid: Sleep Apnea Diagnosis | 27.83 |
| Comorbid: Salicylate sensitive | 27.49 |
| Pain: Eye pain | 27.46 |
| Neurological: Seasonal Affective Disorder (SAD) | 27.39 |
| Comorbid-Mouth: Gingivits / Gum Disease | 27.31 |
| Condition: Generalized anxiety disorder | 27.21 |
| Condition: Post-Traumatic Stress Disorder | 27.09 |
| DePaul University Fatigue Questionnaire : Night sweats | 27.05 |
| Neurological-Sleep: Night Sweats | 26.96 |
| Autonomic Manifestations: exertional dyspnea | 26.96 |
| Comorbid: Constipation and Explosions (not diarrohea) | 26.46 |
| Autonomic Manifestations: cardiac arrhythmias | 26.39 |
| Neurological-Sleep: Vivid Dreams/Nightmares | 26.39 |
| Autism: High Functioning | 26.32 |
| DePaul University Fatigue Questionnaire : Tense muscles | 26.27 |
| Infection: Lyme | 26.26 |
| Comorbid: Mold Sensitivity / Exposure | 26.26 |
| Autonomic Manifestations: Cortisol disorders or irregularity | 26.22 |
| Onset: less than 32 years since onset | 26.14 |
| DePaul University Fatigue Questionnaire : Frequently get words or numbers in the wrong order | 26.05 |
| Pain: Sensitivity to pain | 26.02 |
| Blood Type: B Positive | 25.99 |
| Other: Sensitivity to vibrations | 25.97 |
| Autonomic Manifestations: bladder dysfunction | 25.96 |
| Joint: Stiffness and swelling | 25.95 |
| Comorbid: Raynaud’s syndrome (Skin discoloration) | 25.93 |
| DePaul University Fatigue Questionnaire : Confusion/disorientation | 25.87 |
| DePaul University Fatigue Questionnaire : New trouble with math | 25.87 |
| Immune Manifestations: Thick blood / Hypercoagulation | 25.82 |
| DePaul University Fatigue Questionnaire : Concern with driving | 25.82 |
| DePaul University Fatigue Questionnaire : Weight change | 25.79 |
| Pain: Myofascial pain | 25.77 |
| Comorbid-Mouth: Mouth Sores | 25.76 |
| DePaul University Fatigue Questionnaire : Chilled or shivery | 25.74 |
| Official Diagnosis: Fibromyalgia | 25.73 |
| Neuroendocrine: Feeling like you have a high temperature | 25.65 |
| Neurological-Sleep: Chaotic diurnal sleep rhythms (Erratic Sleep) | 25.54 |
| Neuroendocrine Manifestations: Excessive adrenaline | 25.52 |
| Neurological: fasciculations | 25.50 |
| Age: 60-70 | 25.48 |
| Autonomic: Inability to tolerate an upright position | 25.39 |
| Comorbid: Mood Swings | 25.38 |
| Comorbid: Inflammatory bowel disease | 25.37 |
| Autonomic Manifestations: urinary frequency dysfunction | 25.32 |
| Autonomic: Dizziness or fainting | 25.31 |
| DePaul University Fatigue Questionnaire : Depression | 25.30 |
| Neuroendocrine: Temperature fluctuations throughout the day | 25.29 |
| Comorbid: Restless Leg | 25.29 |
| Comorbid: Snoring (NOT Sleep Apnea | 25.29 |
| DePaul University Fatigue Questionnaire : Nausea | 25.28 |
| Official Diagnosis: Attention deficit hyperactivity disorder (ADHD) | 25.24 |
| DePaul University Fatigue Questionnaire : Feeling like you have a temperature | 25.22 |
| Infection: Human Herpesvirus 6 (HHV6) | 25.22 |
| Neurological: High degree of Empathy before onset | 25.21 |
| DePaul University Fatigue Questionnaire : Chemical sensitivity | 25.19 |
| DePaul University Fatigue Questionnaire : Sore Throat | 25.17 |
| Neuroendocrine Manifestations: abnormal appetite | 25.17 |
| General: Anhedonia (inability to feel pleasure) | 25.13 |
| Neurological-Vision: Blurred Vision | 25.09 |
| Neurological-Vision: photophobia (Light Sensitivity) | 25.02 |
| Immune Manifestations: tender lymph nodes | 24.97 |
| DePaul University Fatigue Questionnaire : Upset stomach | 24.95 |
| Pain: Aching of the eyes or behind the eyes | 24.86 |
| Neurological: Confusion | 24.86 |
| Neuroendocrine Manifestations: marked weight change | 24.83 |
| DePaul University Fatigue Questionnaire : Dizziness | 24.74 |
| Neuroendocrine Manifestations: subnormal body temperature | 24.68 |
| DePaul University Fatigue Questionnaire : Racing heart | 24.64 |
| Immune: Tender / sore lymph nodes | 24.62 |
| Comorbid: Fibromyalgia | 24.58 |
| Comorbid: Migraine | 24.54 |
| General: Myalgia (pain) | 24.54 |
| Neurocognitive: Feeling disoriented | 24.51 |
| Official Diagnosis: Depression | 24.34 |
| Neurocognitive: Unable to focus vision and/or attention | 24.34 |
| Neurological: Neuropathy | 24.34 |
| DePaul University Fatigue Questionnaire : Abnormal sensitivity to light | 24.32 |
| Physical: Good Air Quality | 24.27 |
| Official Diagnosis: Gastroesophageal reflux disease (GERD) | 24.27 |
| Joint: Tenderness | 24.26 |
| Physical: Tonsils removed | 24.24 |
| Comorbid: Methylation issues (MTHFR) | 24.23 |
| DePaul University Fatigue Questionnaire : Tingling feeling | 24.20 |
| Other: Sensitivity to mold | 24.17 |
| Neurological: emotional overload | 24.17 |
| Official Diagnosis: COVID19 (Fully Recovered) | 24.15 |
| Comorbid-Mouth: Dry Mouth | 24.13 |
| Physical: Steps Per Day 8000-16000 | 24.12 |
| Autonomic: Shortness of breath | 24.04 |
| Neuroendocrine: Lack of appetite | 24.03 |
| DePaul University Fatigue Questionnaire : Slow to react | 23.99 |
| Neuroendocrine Manifestations: loss of adaptability | 23.93 |
| DePaul University Fatigue Questionnaire : Rash | 23.90 |
| Autonomic: Irregular heartbeats | 23.87 |
| Physical: Organic Diet | 23.80 |
| Immune: Viral infections with prolonged recovery periods | 23.79 |
| Autonomic Manifestations: light-headedness | 23.75 |
| DePaul University Fatigue Questionnaire : Trouble expressing thoughts | 23.70 |
| DePaul University Fatigue Questionnaire : Feel unsteady on feet | 23.61 |
| Official Diagnosis: Mast Cell Dysfunction | 23.61 |
| Onset: less than 02 years since onset | 23.60 |
| Neurological: Difficulty reading | 23.59 |
| Neurological: Difficulty processing information (Understanding) | 23.57 |
| Immune Manifestations: Inflammation of skin, eyes or joints | 23.55 |
| Neuroendocrine Manifestations: sweating episodes | 23.42 |
| Neurological: Executive Decision Making (Difficulty making) | 23.39 |
| DePaul University Fatigue Questionnaire : Absent-mindedness | 23.34 |
| General: Heavy feeling in arms and legs | 23.32 |
| Infection: Epstein-Barr virus | 23.30 |
| DePaul University Fatigue Questionnaire : Abdomen pain | 23.29 |
| Neuroendocrine Manifestations: Air Hunger | 23.25 |
| Pain: Joint pain | 23.25 |
| Neurological-Sleep: Inability for deep (delta) sleep | 23.24 |
| Autonomic Manifestations: Orthostatic intolerance | 23.20 |
| DePaul University Fatigue Questionnaire : Does physical activity make you feel better | 23.18 |
| Comorbid: Multiple Chemical Sensitivity | 23.15 |
| Post-exertional malaise: Next-day soreness after everyday activities | 23.13 |
| Official Diagnosis: Autoimmune Disease | 23.09 |
| DePaul University Fatigue Questionnaire : Frequently loose train of thought | 23.09 |
| Immune Manifestations: Mucus in the stool | 23.08 |
| Condition: Non-Celiac Gluten Sensitivity | 23.05 |
| DePaul University Fatigue Questionnaire : Difficulty retaining information | 23.04 |
| DePaul University Fatigue Questionnaire : Slowness of thought | 23.01 |
| Neurological: Dysautonomia | 22.95 |
| DePaul University Fatigue Questionnaire : Difficulty reasoning things out | 22.94 |
| Comorbid-Mouth: TMJ / Dysfunction of the temporomandibular joint syndrome | 22.93 |
| DePaul University Fatigue Questionnaire : Eye pain | 22.92 |
| DePaul University Fatigue Questionnaire : Pain in Multiple Joints without Swelling or Redness | 22.90 |
| Neuroendocrine Manifestations: Painful menstrual periods | 22.90 |
| Neurological: Myoclonic jerks or seizures | 22.89 |
| DePaul University Fatigue Questionnaire : Shortness of breath | 22.88 |
| Autonomic: Ocassional Tachycardia (Rapid heart beat) | 22.86 |
| Immune Manifestations: recurrent flu-like symptoms | 22.86 |
| Neurological: Cognitive/Sensory Overload | 22.84 |
| DePaul University Fatigue Questionnaire : Difficulty following things | 22.84 |
| Autonomic Manifestations: Postural orthostatic tachycardia syndrome (POTS) | 22.83 |
| Immune: Chronic Sinusitis | 22.83 |
| DePaul University Fatigue Questionnaire : Temperature lower than normal | 22.82 |
| DePaul University Fatigue Questionnaire : Impaired Memory & concentration | 22.76 |
| Comorbid: Hypothyroidism | 22.74 |
| DePaul University Fatigue Questionnaire : Hot or Cold spells | 22.72 |
| Official Diagnosis: Autism | 22.72 |
| DePaul University Fatigue Questionnaire : Muscle weakness | 22.68 |
| Neuroendocrine Manifestations: Muscle weakness | 22.64 |
| Autonomic Manifestations: nausea | 22.62 |
| Autonomic: Nausea | 22.59 |
| Age: 30-40 | 22.59 |
| Blood Type: O Positive | 22.58 |
| DePaul University Fatigue Questionnaire : Mood swings | 22.58 |
| Sleep: Need to nap daily | 22.57 |
| Neuroendocrine: Feeling hot or cold for no reason | 22.53 |
| DePaul University Fatigue Questionnaire : Sensitivity to Alcohol | 22.52 |
| DePaul University Fatigue Questionnaire : Blurred Vision | 22.49 |
| Onset: less than 04 years since onset | 22.47 |
| Physical: Amalgam fillings | 22.43 |
| Neuroendocrine Manifestations: Paraesthesia (tingling burning of skin) | 22.42 |
| Post-exertional malaise: Mentally tired after the slightest effort | 22.40 |
| Neurological-Vision: inability to focus eye/vision | 22.38 |
| Immune: Recurrent Sore throat | 22.38 |
| DePaul University Fatigue Questionnaire : Difficulty staying asleep | 22.37 |
| DePaul University Fatigue Questionnaire : Need to have to focus on one thing at a time | 22.37 |
| DePaul University Fatigue Questionnaire : Ringing in the Ears | 22.34 |
| Immune Manifestations: Diarrhea | 22.26 |
| General: Headaches | 22.25 |
| Immune Manifestations: Hair loss | 22.25 |
| DePaul University Fatigue Questionnaire : Difficulty comprehending Information | 22.24 |
| Neuroendocrine Manifestations: Rapid muscular fatiguability | 22.24 |
| Post-exertional malaise: Worsening of symptoms after mild mental activity | 22.24 |
| Physical: Breastfed | 22.23 |
| Physical: Long term (chronic) stress | 22.22 |
| DePaul University Fatigue Questionnaire : Headaches | 22.21 |
| Physical: Steps Per Day 2000-4000 | 22.21 |
| Neuroendocrine: Lost or gained weight without trying | 22.19 |
| Autonomic Manifestations: irritable bowel syndrome | 22.18 |
| Post-exertional malaise: Physically drained or sick after mild activity | 22.15 |
| General: Depression | 22.15 |
| Physical: Pets | 22.14 |
| Neurological: Joint hypermobility | 22.12 |
| DePaul University Fatigue Questionnaire : Forgetting what you are trying to say | 22.10 |
| Neurological-Audio: hypersensitivity to noise | 22.04 |
| Comorbid: High Anxiety | 22.02 |
| Immune Manifestations: Inflammation (General) | 21.97 |
| Age: 50-60 | 21.94 |
| Onset: 2000-2010 | 21.89 |
| Immune: Flu-like symptoms | 21.85 |
| DePaul University Fatigue Questionnaire : Anxiety/tension | 21.78 |
| Sleep: Waking up early in the morning (e.g. 3 AM) | 21.76 |
| Post-exertional malaise: Muscle fatigue after mild physical activity | 21.75 |
| Post-exertional malaise: Physically tired after minimum exercise | 21.70 |
| Comorbid: Constipation and Diarrohea (not explosions) | 21.70 |
| DePaul University Fatigue Questionnaire : Muscle Pain (i.e., sensations of pain or aching in your muscles. This does not include weakness or pain in other areas such as joints) | 21.69 |
| Comorbid-Mouth: Bruxism – Jaw cleanching / Teeth grinding | 21.65 |
| Condition: Acne | 21.63 |
| Immune: Sensitivity to smell/food/medication/chemicals | 21.61 |
| Blood Type: A Positive | 21.61 |
| Immune Manifestations: Abdominal Pain | 21.59 |
| Physical: Northern European | 21.58 |
| Pain: Pain or aching in muscles | 21.54 |
| Neuroendocrine Manifestations: intolerance of extremes of heat and cold | 21.52 |
| General: Sinus issues with headaches | 21.48 |
| Post-exertional malaise: Post-exertional malaise | 21.48 |
| Post-exertional malaise: Rapid muscular fatigability, | 21.44 |
| Neuroendocrine: Cold limbs (e.g. arms, legs hands) | 21.42 |
| Immune Manifestations: Chronic Flatus / Flatulence / gas | 21.39 |
| DePaul University Fatigue Questionnaire : Allergies | 21.39 |
| DePaul University Fatigue Questionnaire : Does physical activity make you feel worse | 21.37 |
| Onset: less than 16 years since onset | 21.35 |
| Neuroendocrine Manifestations: Dry Eye (Sicca or Sjogren Syndrome) | 21.35 |
| Neurocognitive: Slowness of thought | 21.35 |
| Neurological: Word-finding problems | 21.30 |
| Neurological: Impairment of concentration | 21.29 |
| Immune Manifestations: new food sensitivities | 21.28 |
| Post-exertional malaise: Worsening of symptoms after mild physical activity | 21.27 |
| Physical: Steps Per Day 4000-8000 | 21.26 |
| Immune Manifestations: Alcohol Intolerant | 21.25 |
| Physical: Steps Per Day < 2000 | 21.24 |
| Condition: ME/CFS without IBS | 21.23 |
| DePaul University Fatigue Questionnaire : Need to nap during each day | 21.23 |
| DePaul University Fatigue Questionnaire : Poor Appetite | 21.23 |
| DePaul University Fatigue Questionnaire : Fatigue | 21.21 |
| Physical: Eastern European | 21.21 |
| Neurocognitive: Can only focus on one thing at a time | 21.20 |
| Condition: ME/CFS with IBS | 21.15 |
| Neurocognitive: Problems remembering things | 21.13 |
| DePaul University Fatigue Questionnaire : Difficulty recalling information | 21.10 |
| DePaul University Fatigue Questionnaire : Difficulty falling asleep | 21.08 |
| Sleep: Daytime drowsiness | 21.07 |
| Neurocognitive: Absent-mindedness or forgetfulness | 21.07 |
| Post-exertional malaise: Rapid cognitive fatigability, | 21.05 |
| Onset: Sudden | 21.03 |
| DePaul University Fatigue Questionnaire : Easily irritated | 21.03 |
| DePaul University Fatigue Questionnaire : Walking up early in the morning (e.g. 3AM) | 21.03 |
| Immune Manifestations: medication sensitivities. | 21.01 |
| Neurological-Audio: Tinnitus (ringing in ear) | 20.98 |
| Comorbid: Small intestinal bacterial overgrowth (SIBO) | 20.96 |
| Physical: Work-Sitting | 20.96 |
| Neuroendocrine: Alcohol intolerance | 20.91 |
| Neurological-Sleep: Insomnia | 20.91 |
| Onset: Gradual | 20.90 |
| Neurocognitive: Difficulty understanding things | 20.84 |
| Autonomic Manifestations: palpitations | 20.80 |
| Neurological: Short-term memory issues | 20.71 |
| DePaul University Fatigue Questionnaire : Post-exertional malaise, feeling worse after doing activities that require either physical or mental exertion | 20.52 |
| Autonomic: Heart rate increase after standing | 20.51 |
| Immune Manifestations: Bloating | 20.51 |
| DePaul University Fatigue Questionnaire : Unrefreshing Sleep, that is waking up feeling tired | 20.49 |
| DePaul University Fatigue Questionnaire : Difficulty finding the right word | 20.46 |
| Comorbid: Histamine or Mast Cell issues | 20.41 |
| Gender: Female | 20.35 |
| Post-exertional malaise: Inappropriate loss of physical and mental stamina, | 20.33 |
| Neurocognitive: Difficulty expressing thoughts | 20.27 |
| Post-exertional malaise: Difficulty reading after mild physical or mental activity | 20.25 |
| Official Diagnosis: Chronic Fatigue Syndrome (CFS/ME) | 20.20 |
| Post-exertional malaise: General | 20.19 |
| Official Diagnosis: COVID19 (Long Hauler) | 20.18 |
| Official Diagnosis: Allergic Rhinitis (Hay Fever) | 20.06 |
| Neuroendocrine Manifestations: Poor gut motility | 20.04 |
| Sleep: Problems staying asleep | 20.03 |
| Onset: 2010-2020 | 20.01 |
| Neuroendocrine Manifestations: worsening of symptoms with stress. | 19.94 |
| Neurocognitive: Brain Fog | 19.88 |
| Immune Manifestations: general malaise | 19.84 |
| Sleep: Problems falling asleep | 19.75 |
| Neuroendocrine Manifestations: cold extremities | 19.68 |
| Official Diagnosis: Irritable Bowel Syndrome | 19.64 |
| Neurocognitive: Difficulty paying attention for a long period of time | 19.57 |
| Immune Manifestations: Constipation | 19.42 |
| Age: 20-30 | 19.37 |
| Age: 40-50 | 18.92 |
| Sleep: Unrefreshed sleep | 18.84 |
| General: Fatigue | 18.39 |
| Gender: Male | 18.29 |






Recent Comments