Sue, a reader in Australia, shared her experience below on the challenge of taking supplements. Other people may have the same challenge. In some cases, retail products additives can be the source of problems. In this house, we tend to make our own or use additive free.
The problem began when Ken Lassesen’s brilliant AI came up with oils my daughter must take by swallowing them, with high probability of success. Coriander, thyme, lavender, perilla oils. She’s been sick to the point of house-bound for years, and we’ve got to get her into life. She has autism and chronic fatigue, and since 2018 Ken Lassesen’s AI has at least got her out of bed (unexpectedly, skads of licorice and thyme leaves are key players here) when no kindly, well-informed, hard-working doctor could achieve that.
So- where to get the oils? First call was to my excellent local pharmacy in Sydney – Newton’s – and they told me that their oils shouldn’t be ingested, but they’d heard of a place “somewhere in NSW” called “something like” TERRA, and they were “a bit expensive”. I found and rang TERRA, and they sent me the oils, one expensive from a company that’s licensed to provide ingestible oils, the other from a second company that I was assured was as good but hasn’t gone through the rigours of getting a license.
Now, how to get them into her. We tried dripping it into Bonvit gel caps from our local chemist with some “blotting paper” in the bottom of thiamine which she has to take anyway, but they disintegrated almost as we looked at them. Bonvit are fine with powders, but oils aren’t powders! Then we tried Surgipack’s capsules which were sturdier so they lasted until she put them in her mouth. A few seconds later, howls of pain.
Next morning we thought we had the solution and tracked down two sizes of Surgipack, so we put the oil and thiamine mixture into the smaller one and put that inside the bigger one, carefully wiping down the outside surface. We were very pleased with ourselves until a phone call with howls of pain with a burning oesophagus and stomach. I talked her into trying to take them right in the middle of breakfast, and she agreed to give them another go. It was still painful afterwards but she bravely soldiered on, saying that “it might be working” ! But after five days, the pain got too much to bear and she said she’d “never have that stuff again. However good it’s supposed to be”.
But there must be nasty meds taken all the time, so how do the commercial companies get them into people without the whole nation howling in pain, with mass revolts? They must know something we don’t.
We have a lot of private compounding chemists here so I rang around and asked them, one after another, if they used special capsules and could I buy some. But they make their money out of compounding, not selling their ingredients, so no. At last I chanced on a chatty girl who said what I needed was enteric-coated capsules”, but that her company couldn’t supply them. She vaguely mentioned legal reasons.
So then, the internet. We immediately found a supplier of enteric capsules in Australia, The Capsule Guy, costing $17 for 250 capsules. They come in sizes and we chose smallish ones, so we could put them inside a larger Surgipack capsule in case of dribbles.
We began 4 days ago. No howls. Then joy. Yesterday afternoon, her birthday, she went out to the party of a childhood friend who has the same birthday. She only stayed 3 hours before she wilted, but she went out and we are over the moon. Thank you, Ken. You are bringing our daughter into life.
Post-Script
Reading an account like this makes all of the hours that I spent on the blog and web site worthwhile. Thank you Sue for sharing! P.S. Sue started in 2018, it is not an overnight turnaround, it is a slow long march. Each person is unique, as is their microbiome. Microbiome Prescription is specific to an individual’s microbiome and not their diagnosis.

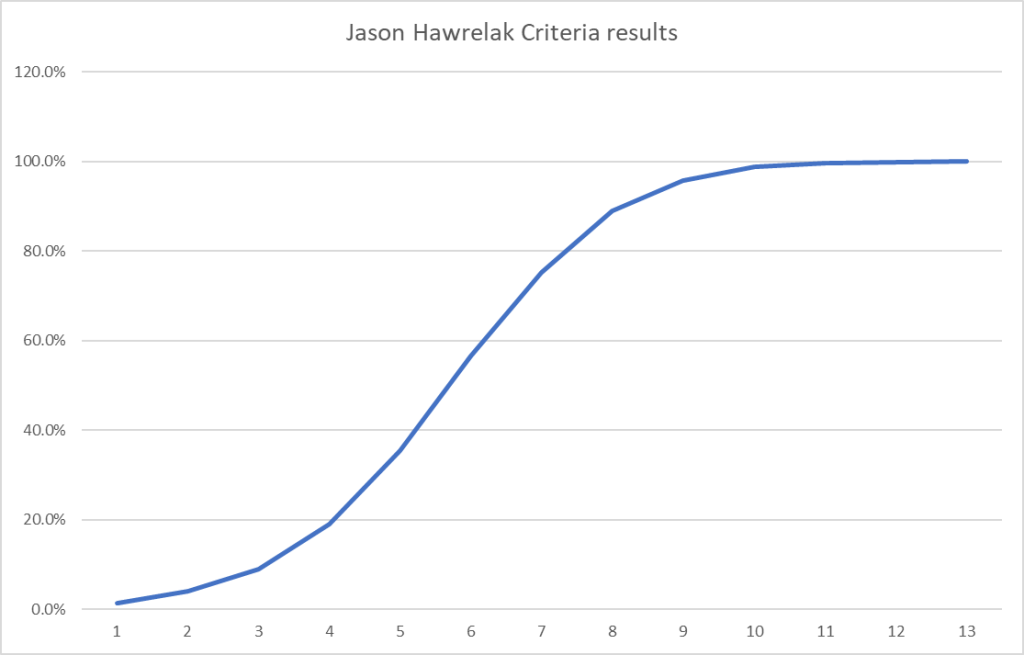
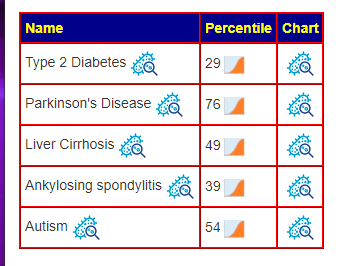
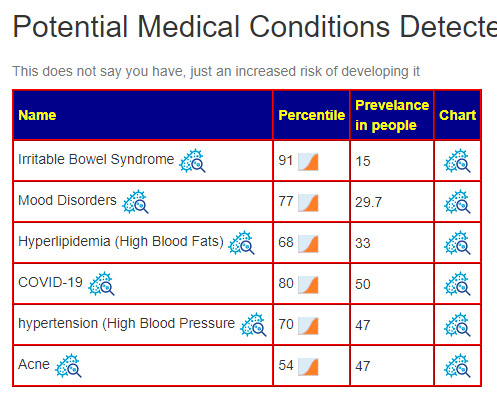
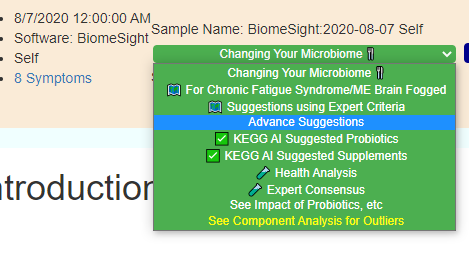


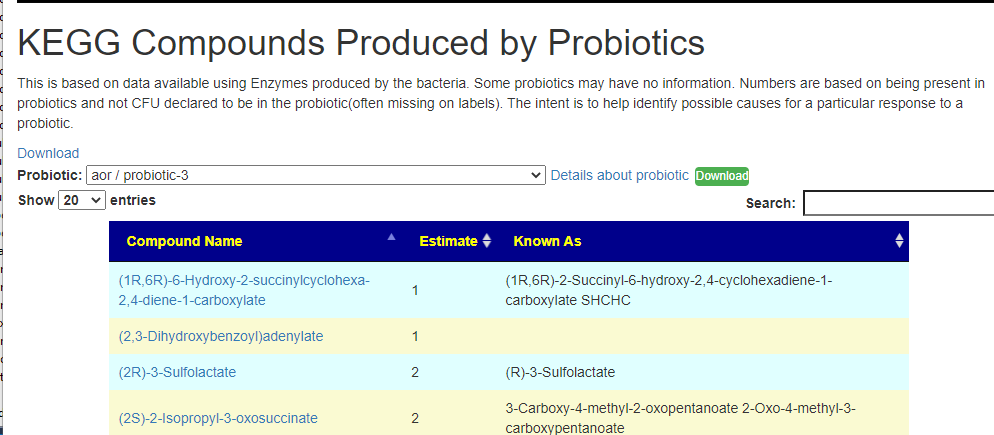
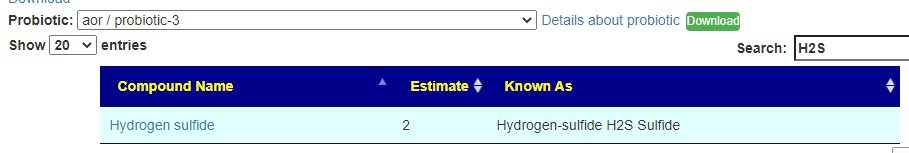
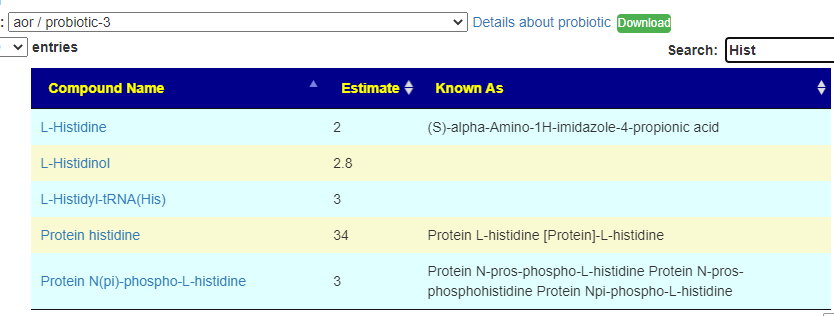


Recent Comments