We have a microbiome, we have lab results, we have official conditions (ICD), we have symptoms. Last, we have substances (for example, probiotics) that modify the microbiome and thus may alter:
- lab results
- official condition status (i.e. mild, severe, acute)
- microbiome
- symptoms (one symptom may disappear or appear)
Information on expected impact of the above come from medical studies.
The typical question is “What should I take to improve {lab results|symptoms|official diagnosis|microbiome}?” The response should be typically, “Base on study A,B,C,K, you should take X to improve {lab results|symptoms|official diagnosis|microbiome}? “
The answers may come indirectly and may be by inference. For example:
I wish to improve my diabetes.
- Severity of diabetes is connected with high A bacteria and low B bacteria and high levels of TNF-alpha
- Substance X has no published studies for diabetes
- Substance X has published studies for decreasing A and not altering B.
- Substance Y has published studies for increasing B and not altering A but it does reduce TNF-alpha levels.
The inference is that you should consider taking X and Y to improve your diabetes. In some cases, you may find something like:
I wish to improve my mother’s Alzheimer’s Disease.
- Severity of Alzheimer’s Disease is connected with high A bacteria and low B bacteria.
- Substance X and Y has published studies for Alzheimer’s Disease showing positive results
- Substance X has published studies for decreasing A and not altering B.
- Substance Y has published studies for increasing B and not altering A.
The database schema below attempts to capture this information from citations (studies).
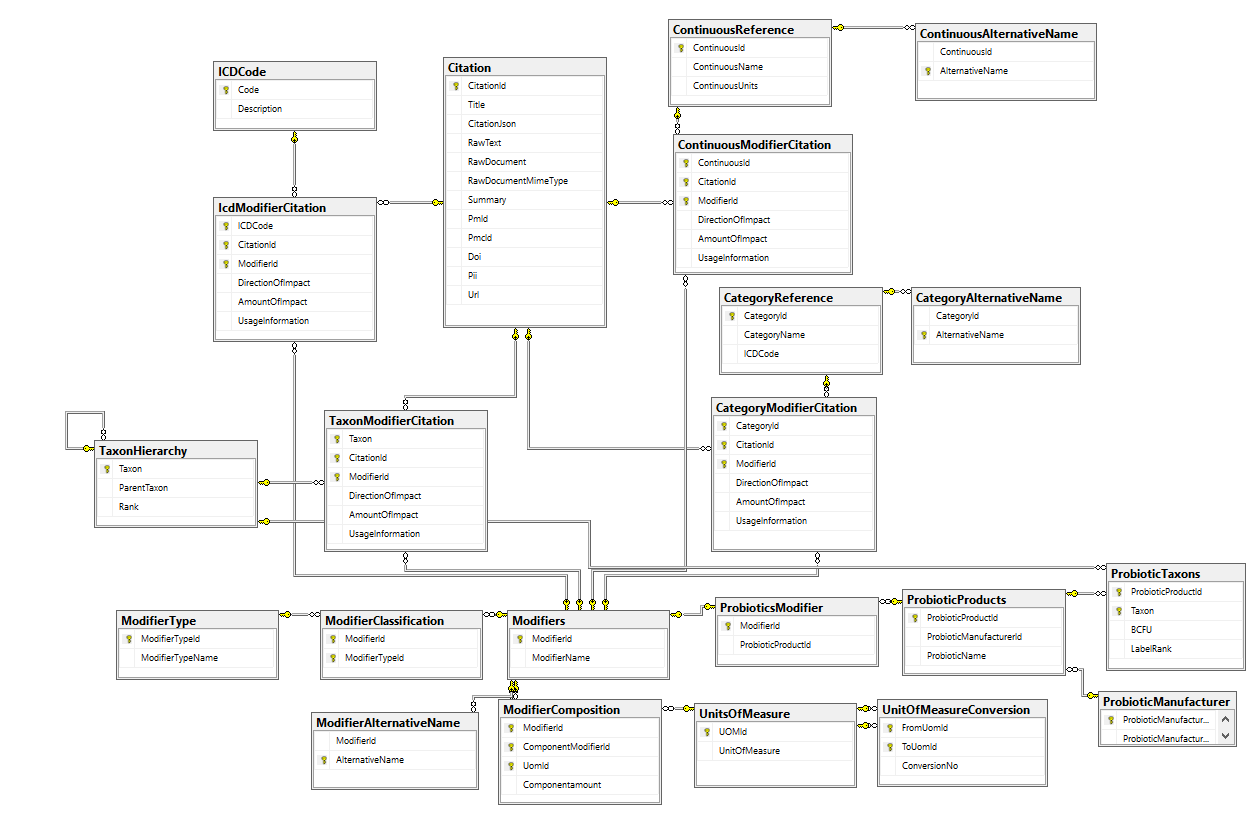
Let us look at what information may be in a study and map the information to tables (following are made up study results for illustrations)
- Salted Herrings at 20gm/day improves IBS from Study A
- Modifier: Salted Herring
- Citation: A
- ICDCode: IBS
- ICDModifierCitation
- DirectionOfImpact: +1
- AmountOfImpact: NULL — nothing reported
- UsageInformation: 20gm/day
- Same study found TNF-Alpha Increases by 20% above control
- Confinuous Reference: TNF-Alpha
- ContinousModifierCitation:
- DirectionOfInpact: +1
- Amount of Impact: 1.2 (1 being no change)
- UsageInformation: 20gm/day
- Same study found Asthma Disappear in 30% of patients
- CategoryReference: Asthma (Yes or No remember)
- ContinousModifierCitation:DirectionOfInpact: -1
- Amount of Impact: 0.8 (1 being no change)
- UsageInformation: 20gm/day
- CategoryReference: Asthma (Yes or No remember)
- Same study found Sillium bacteria increased in patients
- TaxonHierarchy: Sillium
- TaxonModifierCitation:DirectionOfInpact: +1
- Amount of Impact: nothing reported
- UsageInformation: 20gm/day
- TaxonHierarchy: Sillium
So the results of one study ended up with entries in 4 tables.
We have a lot of possible inferences here:
- Sillium impacts TNF-Alpha
- Low Sillium may be associated with Asthma
All of this stuff becomes facts in our Artificial Intelligence/Expert System engine which I will cover in a few weeks.
Alternative Names
Alternative names is actually critical for text mining (i.e. having programs determine if there is important data is a study, paragraph or sentence). Studies may use a multitude of names for the same thing. For example, you may decided to use the latin name for herbs, Hypericum perforatum and then have the alternative names “St. John Wart” and “Saint John Wart”. The alternative names should be unique, hence the unique index is placed on this column.
Bottom Line
Above is the full solution, I have only partially implemented it and the only one of the table that I have been populating has been TaxonModifierCitation. Readers have asked question about TNF-Alpha, Interleukin 10 (IL-10), also known as human cytokine synthesis inhibitory factor (CSIF). My own resources could only stretched to review and processing this table. Ideally, a crowd-source efforts (or a wealthy patron to have Ph.D. students) would allow the full solution to be populated.
Recent Comments