Back Story
I have severe /very severe ME/CFS and have noticed partially dramatic changes (although short lived) when taking a probiotic, especially Miyarisan[Clostridium butyricum].
Analysis
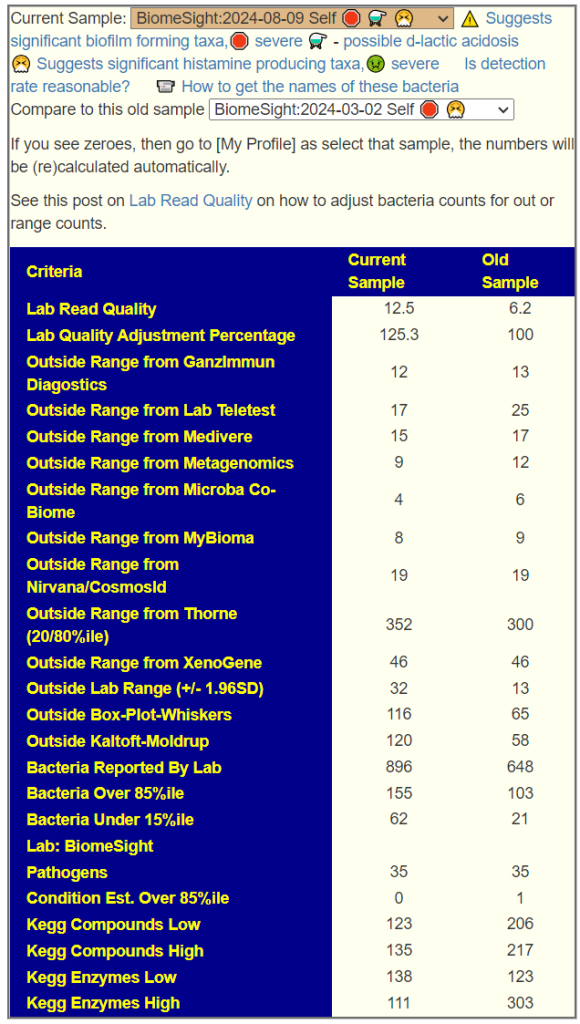
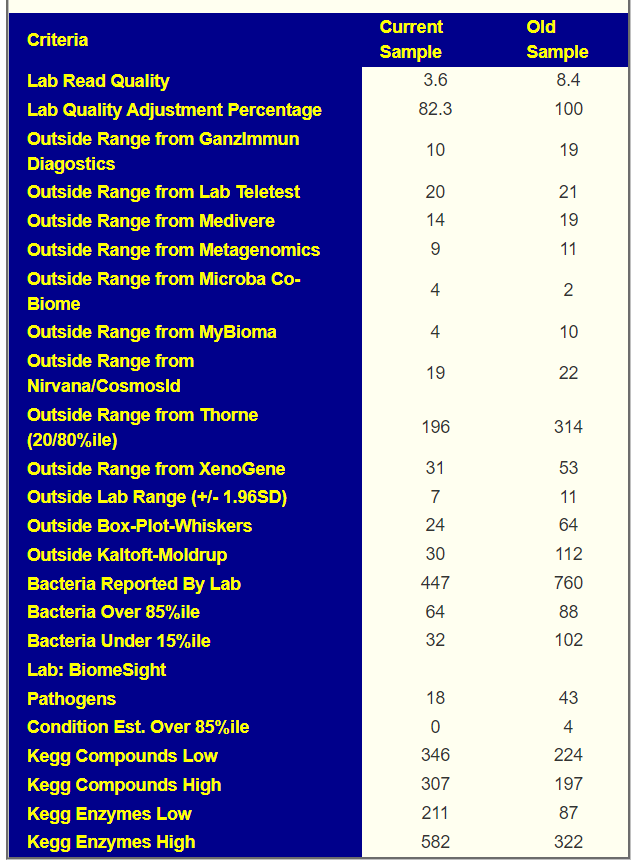
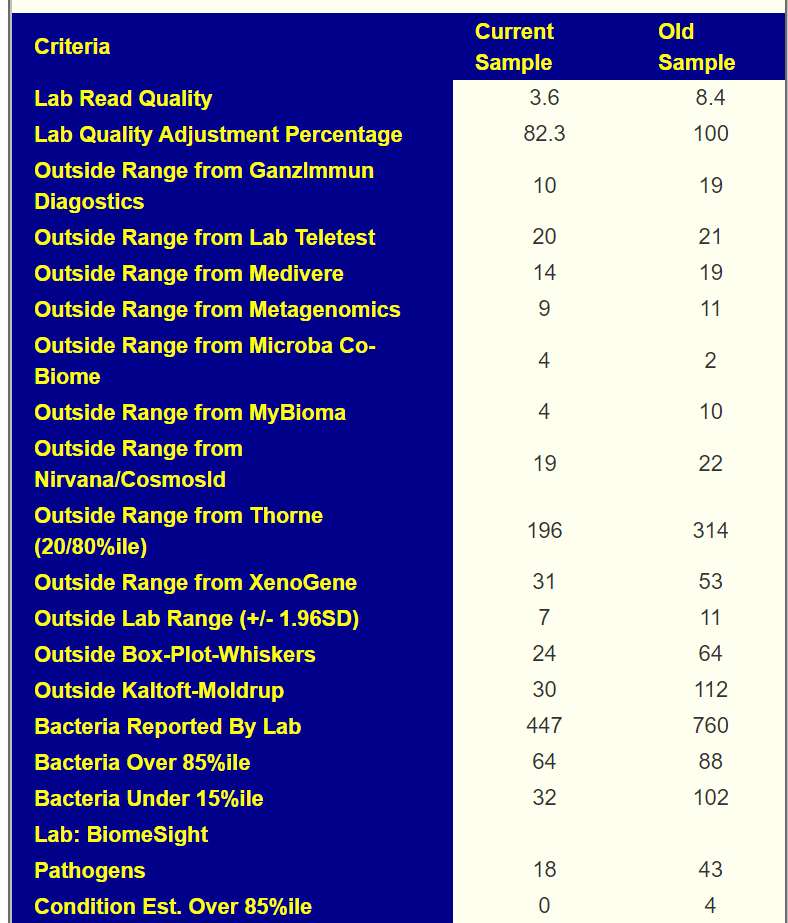
Sample Comparison
My general impression is that this person has lost some ground in terms of reference ranges(more found at extremes), but has gained ground with Kegg Compounds and Enzymes (less ones at extremes).
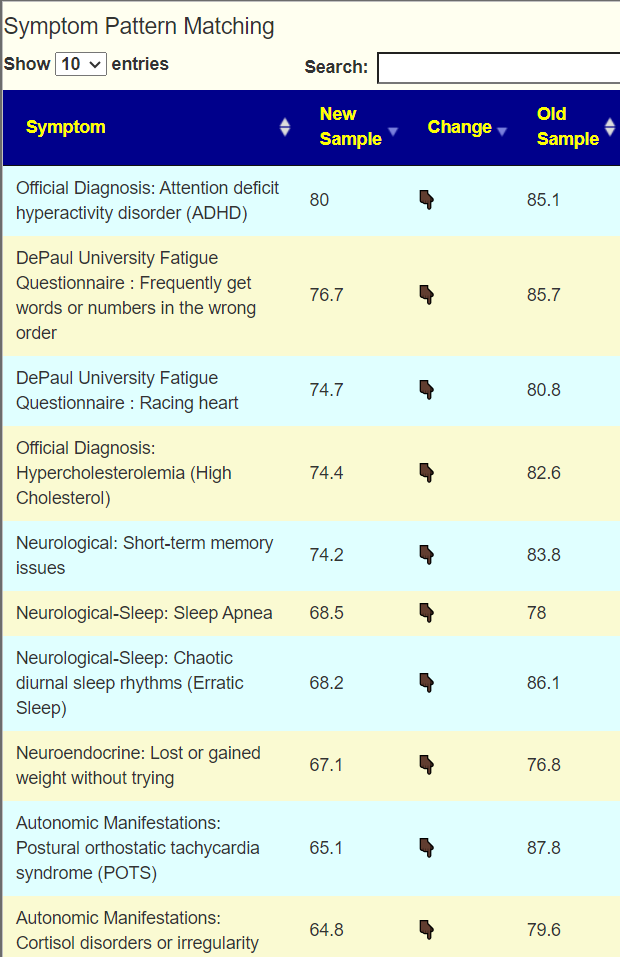
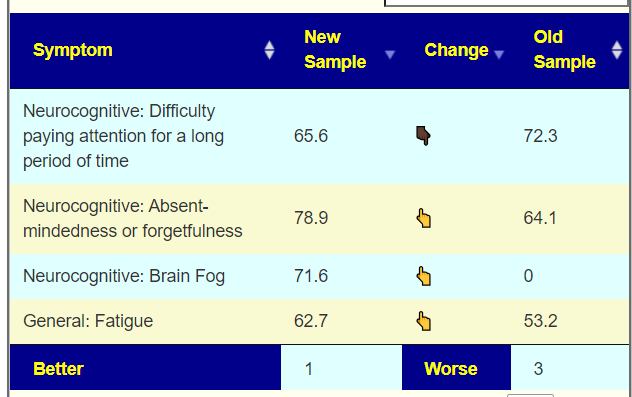
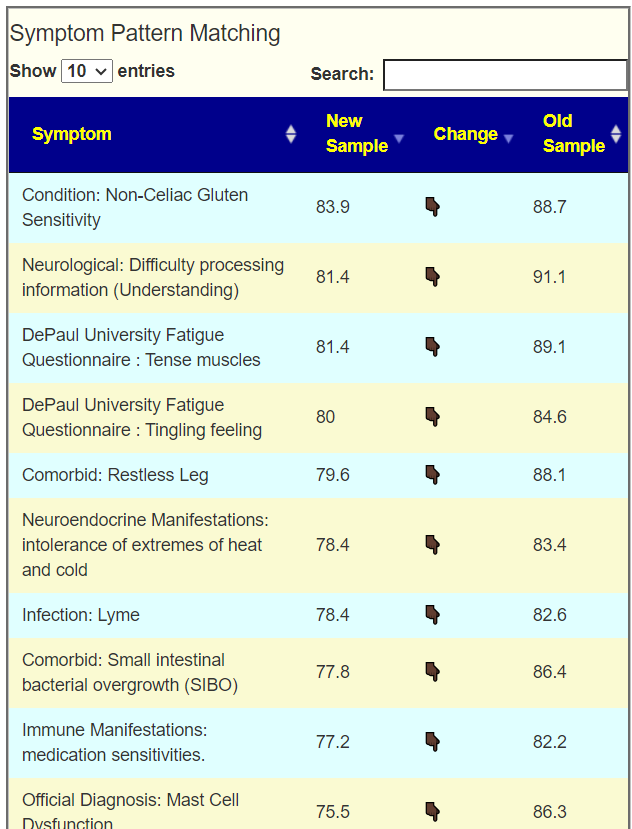
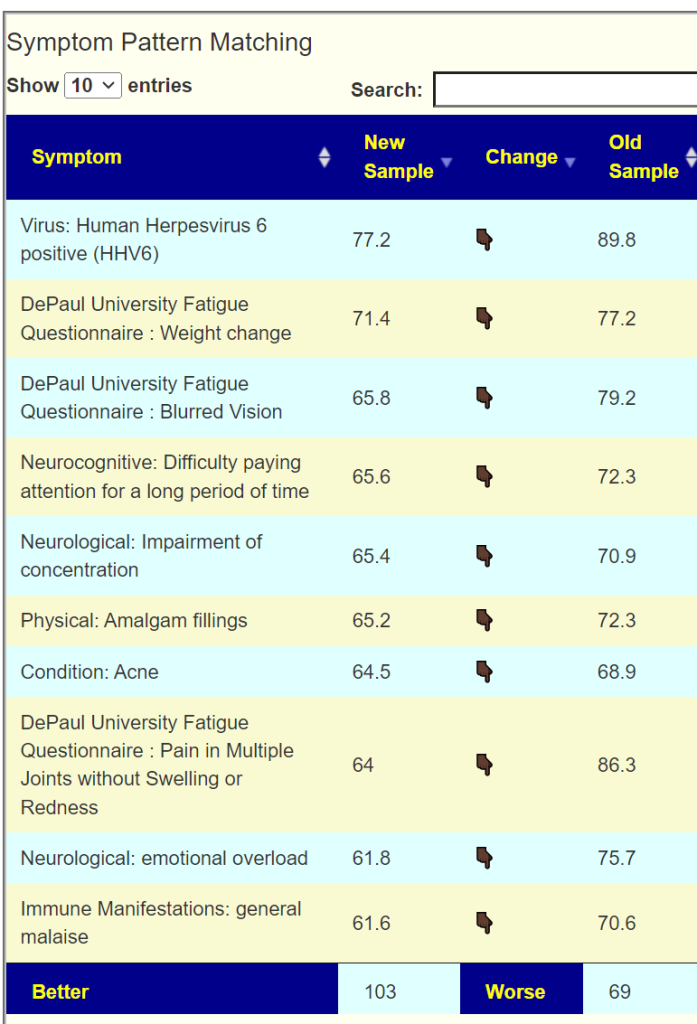
To get better insights, I added a Pattern Matching Comparison. Only symptoms marked in either samples are compared. We see some improvement happened.
Going Forward
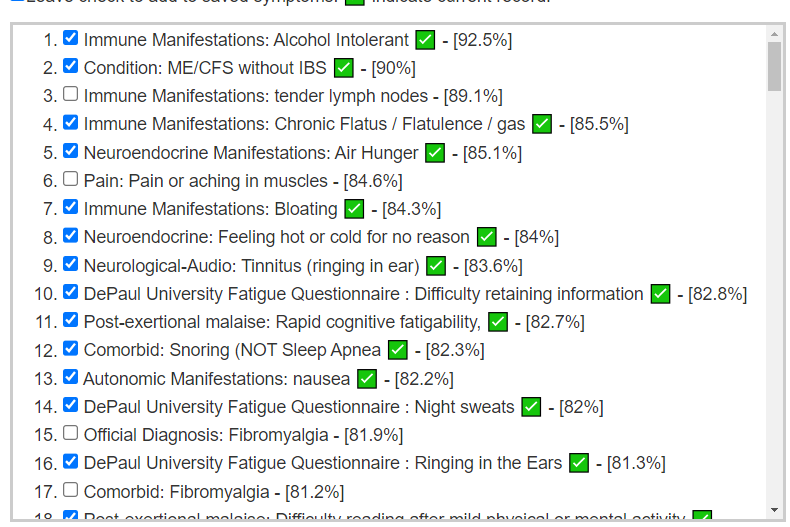
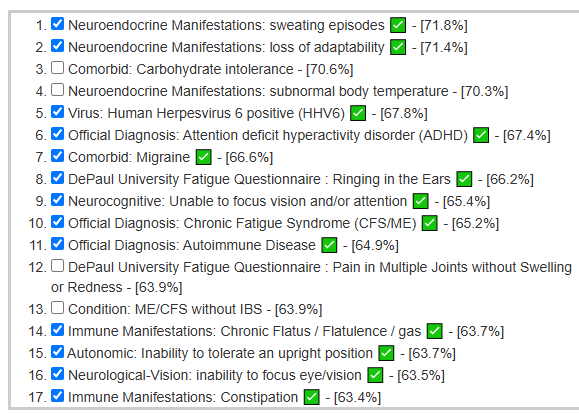
My updated starting point with the new UI when the person has one or more conditions picked to [Beginner-Symptoms: Select bacteria connected with symptoms]. As shown below, we have a large number of symptoms matching the patterns from our data analysis. This suggests that we are likely to pick the right bacteria to focus on (based on statistical evidence – which any skilled person can reproduce using data on https://citizenscience.microbiomeprescription.com/).
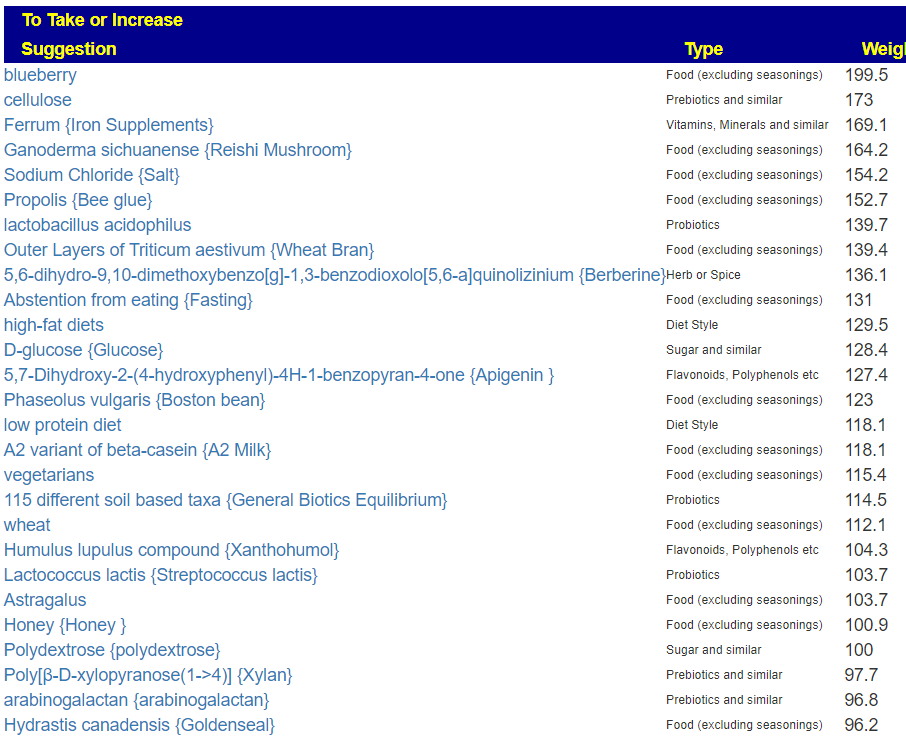
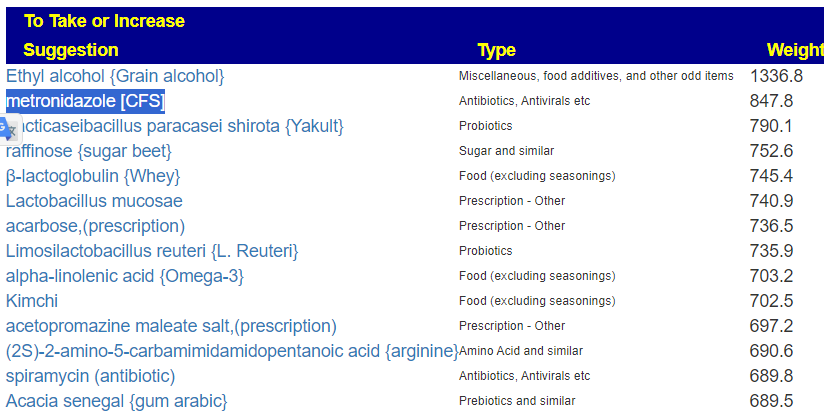
The top suggestions are below

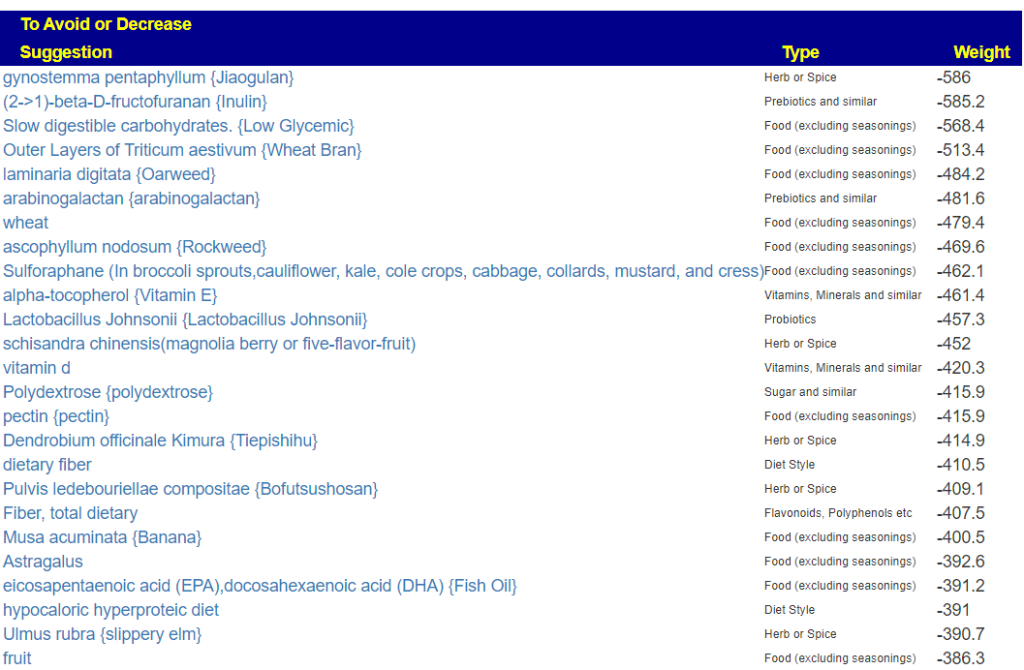
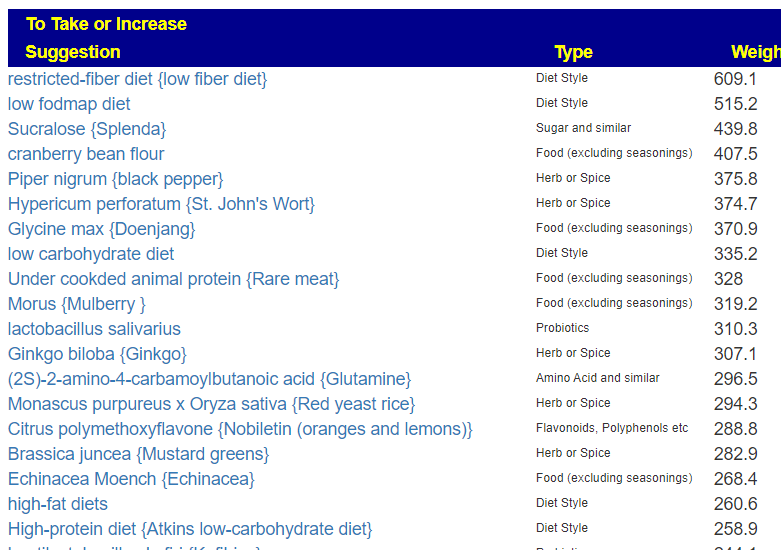
As well as the top avoids
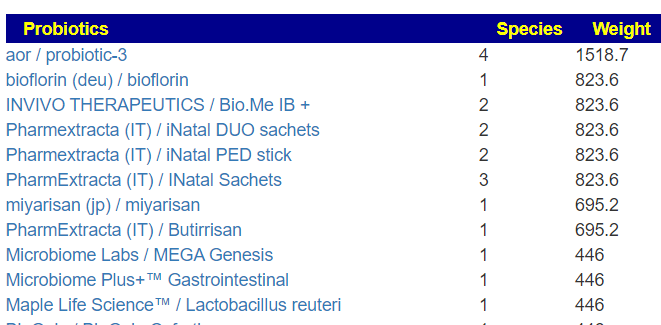
Probiotics
The top probiotics using published studies on PubMed were:
- Lactobacillus gasseri {L.gasseri}
- Lactobacillus reuteri
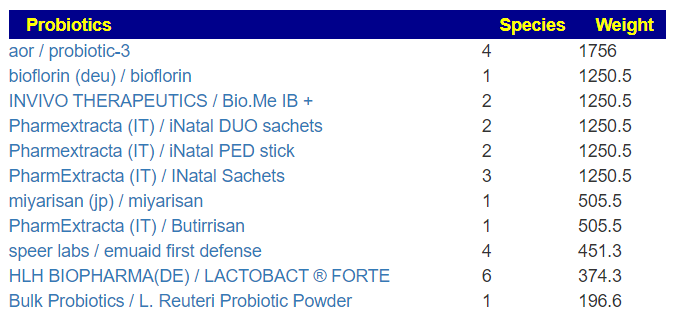
- aor / probiotic-3 (contains miyarisan probiotic)
- miyarisan (jp) / miyarisan
With the new UI, we also have probiotics computed from RNA/DNA of your microbiome and probiotics. Usually I select only low compounds (i.e. some bacteria will be inhibited from starvation).
As is typical with ME/CFS, the top ones are E.Coli probiotics.
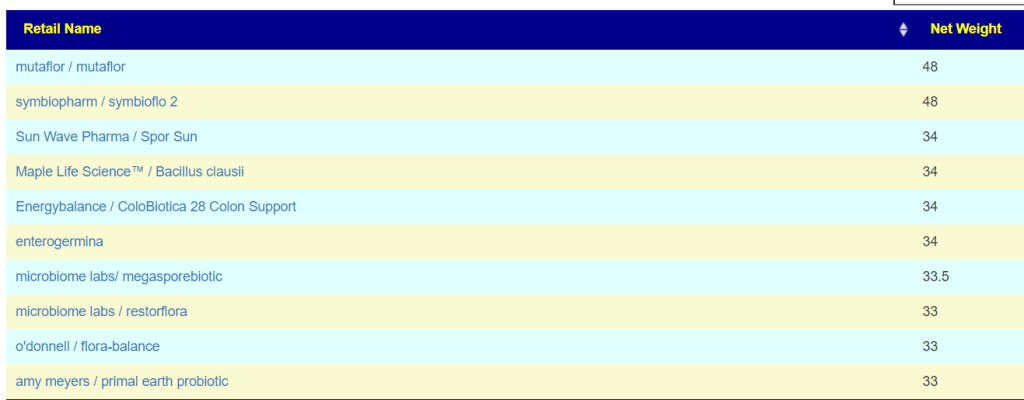
I checked each of the PubMed suggestion to see their relative impact and put in [ ] below.
- Lactobacillus gasseri {L.gasseri} [14]
- Lactobacillus reuteri [22]
- aor / probiotic-3 (contains miyarisan probiotic) [28]
- miyarisan (jp) / miyarisan [24] a.k.a. Clostridium butyricum
I would start with aor / probiotic-3, then one of the Lactobacillus reuteri , then Lactobacillus gasseri and end with miyarisan. While aor and miyarisan both contain the same bacteria species, they are different strains.
- Food avoid list is high in food containing fiber (in agreement with diet style)
Update using new Simple UI
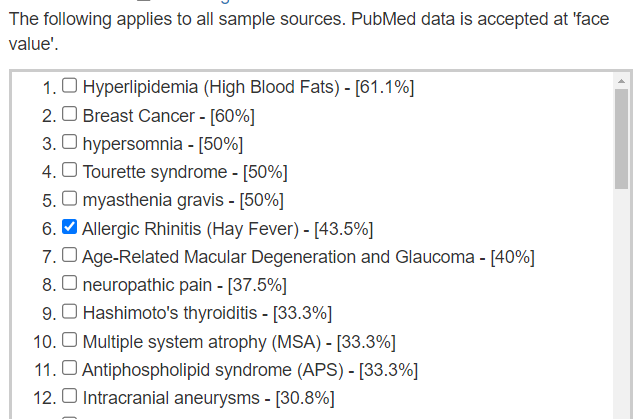
We see our forecast symptoms being accurate very often as shown below, so I did [Beginner-Symptoms: Select bacteria connected with symptoms]. The intent is to focus solely on the bacteria likely causing the symptoms. There were 141 symptoms associations use which resulted in just 50 bacteria being picked. Often the same group of bacteria will cause multiple symptoms (depending on a person’s DNA etc).
The result was dominated by antibiotics and other off-label prescription items (needing a cooperative MD).

Swinging to Probiotics — the usual starting point for many people. The top choices were:
- Lacticaseibacillus casei {L. casei} [one source]
- Lacticaseibacillus paracasei {L.paracasei} – Yakult is the easiest one to get
- Lentilactobacillus kefiri {Kefibios} – Available only in Italy
- Lactobacillus gasseri {L.gasseri} [one source]
These are based on probiotics that has had clinical studies done on them. In other words, those likely to inhibit or encourage desired bacteria shifts. An alternative approach to look for probiotics that produce metabolites and enzymes that the person appears to be low on. The goal is reduce the dysbiosis caused by starvation. The top suggestions are:
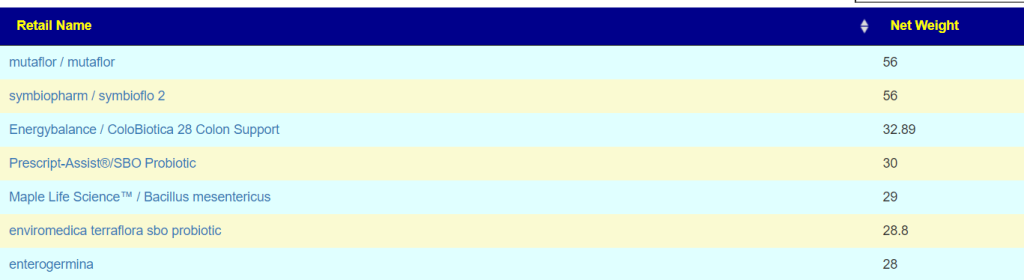
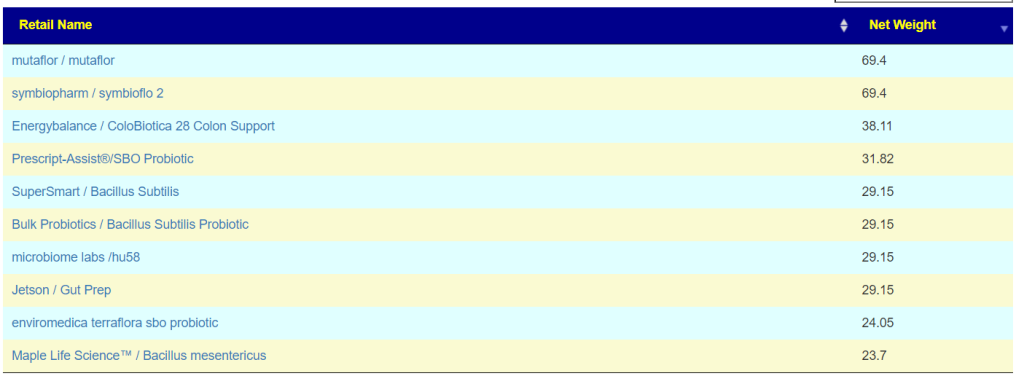
- Escherichia coli – Mutaflor, Symbioflor2 [48 / 103] and on the to take list above.
- Bacillus clausii [34 / 72]
- Bacillus subtilis [32 / 68]
- Lacticaseibacillus casei [25 /54]
- Lacticaseibacillus paracasei [25 / 55]
- Lactobacillus gasseri [14 / 38]
The safest trinity of probiotics is: Escherichia coli, Lacticaseibacillus casei, and Lacticaseibacillus paracasei. Taking each for 1-2 weeks and then rotate to the next. With Lactobacillus gasseri and the two bacillus being worth an experiment afterwards
Important Note: The reader updated their symptoms and this is with the updated symptoms. Changing the selection of bacteria will usually cause shifts of suggestions (see this post)
Bottom Line
The user report of improvement with miyarisan and the suggestions are a nice agreement to see. The issue of being short termed is not atypical to see when there is no rotation of probiotics and antibiotics.
IMHO, probiotics should be viewed as natural antibiotics. As with all antibiotics, antibiotic resistance (probiotic resistance) may developed from continuous use. For Lactobacillus reuteri we have Reuterin; for Clostridium butyricum we have: CBP22, Butyricin 7423, Butyricum M588, Perfringocin 1105. (see Effects of Clostridium butyricum as an Antibiotic Alternative [2023]).
The same applies to herbs and spices with antibiotic characteristics… resistance will often develop from continuous use.
Postscript and Reminder
As a statistician with relevant degrees and professional memberships, I present data and statistical models for evaluation by medical professionals. I am not a licensed medical practitioner and must adhere to strict laws regarding the appearance of practicing medicine. My work focuses on academic models and scientific language, particularly statistics. I cannot provide direct medical advice or tell individuals what to take or avoid.My analyses aim to inform about items that statistically show better odds of improving the microbiome. All suggestions should be reviewed by a qualified medical professional before implementation. The information provided describes my logic and thinking and is not intended as personal medical advice. Always consult with your knowledgeable healthcare provider.
Implementation Strategies
- Rotate bacteria inhibitors (antibiotics, herbs, probiotics) every 1-2 weeks
- Some herbs/spices are compatible with probiotics (e.g., Wormwood with Bifidobacteria)
- Verify dosages against reliable sources or research studies, not commercial product labels. This Dosages page may help.
- There are 3 suppliers of probiotics that I prefer: Custom Probiotics , Maple Life Science™, Bulk Probiotics: see Probiotics post for why
- My preferred provider for herbs etc is Maple Life Science™ – they are all organic, fresh, without fillers, and very reasonably priced.
Professional Medical Review Recommended
Individual health conditions may make some suggestions inappropriate. Mind Mood Microbes outlines some of what her consultation service considers:
A comprehensive medical assessment should consider:
- Terrain-related data
- Signs of low stomach acid, pancreatic function, bile production, etc.
- Detailed health history
- Specific symptom characteristics (e.g., type and location of bloating)
- Potential underlying conditions (e.g., H-pylori, carbohydrate digestion issues)
- Individual susceptibility to specific probiotics
- Nature of symptoms (e.g., headache type – pressure, cluster, or migraine)
- Possible histamine issues
- Colon acidity levels
- SCFA production and acidification needs
A knowledgeable medical professional can help tailor recommendations to your specific health needs and conditions.



Recent Comments