It is one of the factors. Diet is also a factor. Today, there was some sweet studies published on Nature. A few quotes are below.
Variants at the LCT locus associated with Bifidobacterium and other taxa, but they differed according to dairy intake. Furthermore, levels of Faecalicatena lactaris associated with ABO, and suggested preferential utilization of secreted blood antigens as energy source in the gut. Enterococcus faecalis levels associated with variants in the MED13L locus, which has been linked to colorectal cancer. Mendelian randomization analysis indicated a potential causal effect of Morganella on major depressive disorder, consistent with observational incident disease analysis
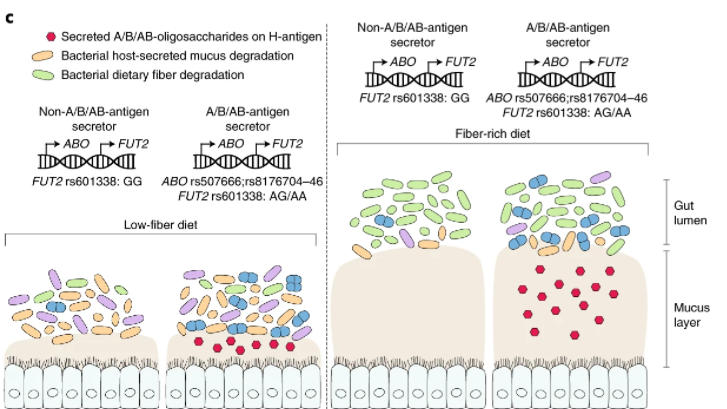
Our associations of F. lactaris (P = 1.10 × 10−12) and Collinsella (P = 2.59 × 10−8) with ABO suggest a possible metabolic link with blood antigens.
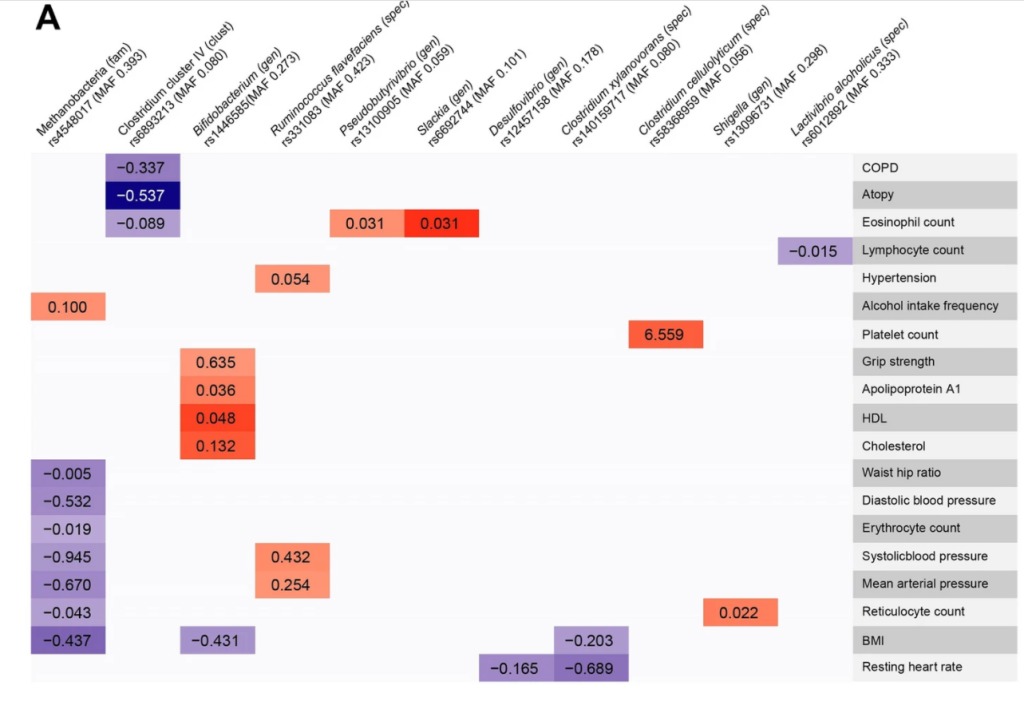
In our study, as in previous work3,5,6,10, the association of LCT variants with Actinobacteria, more specifically Bifidobacterium, is by far the most statistically significant
Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort [2022]
This is well illustrated by this chart in the above study
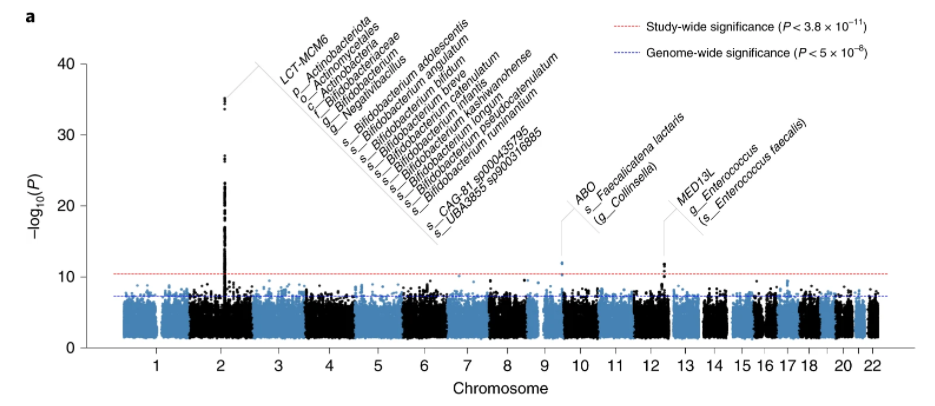
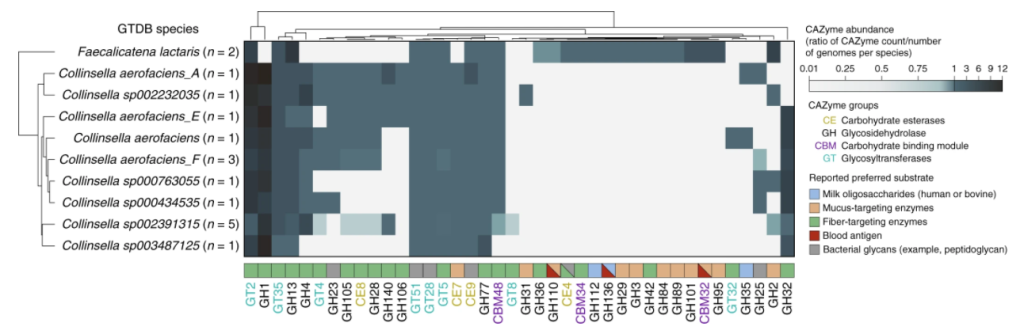
This study illustrate how the presence or absence of some bacteria species depends on the individual’s DNA as shown in the chart below.
They went onwards and show how diet compounds matter more.
This information has been hinted at in earlier studies:
- “association of a functional LCT SNP with the Bifidobacterium genus (P = 3.45 × 10-8) and provide evidence of a gene-diet interaction in the regulation of Bifidobacterium abundance. ” [2016]
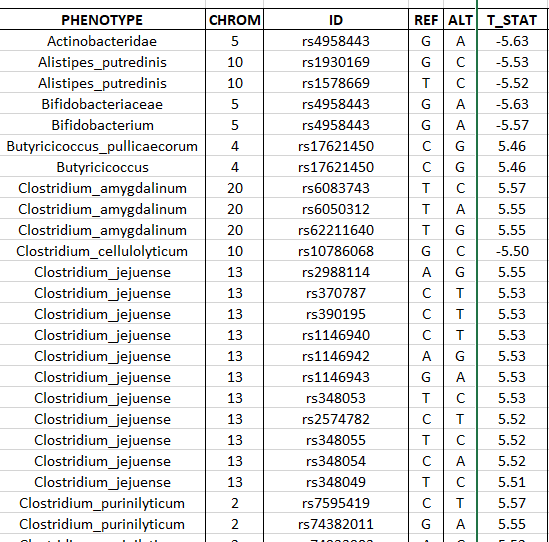
A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals [2019]
We are still in early days, in some cases you may be able to find studies on PubMed by searching for “SNP {bacteria name}”, for example: “SNP clostridium cellulolyticum” or a google search on similar terms. From which I found:
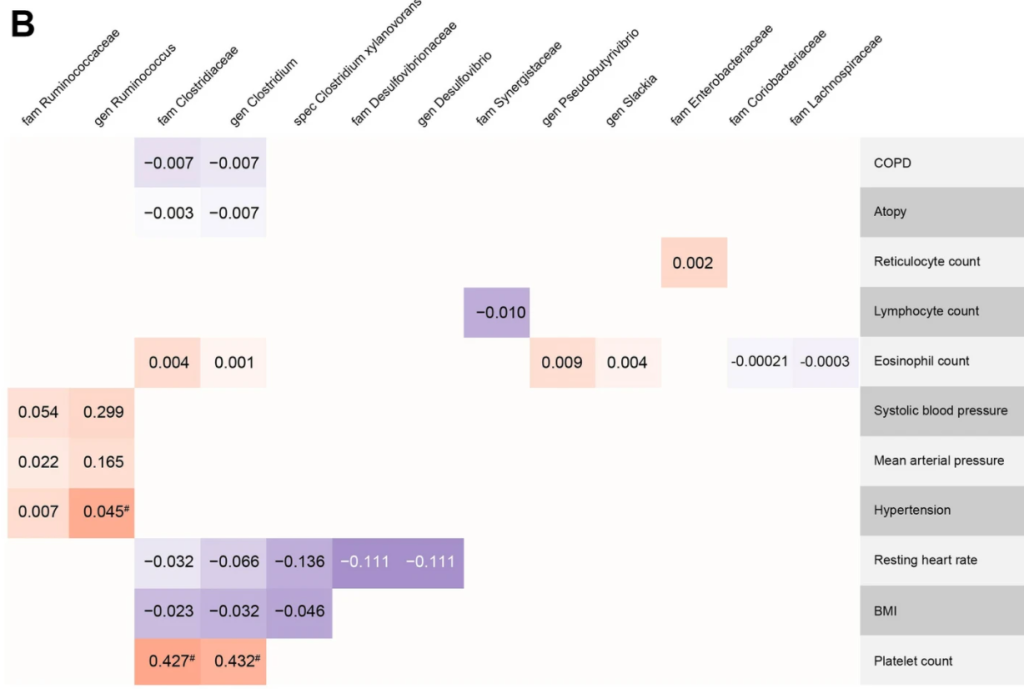
Some charts from this study are shown below:
Recent Comments