A reader asked the following questions:
1. How does the algorithm/tech learn and train? (I am a biologist and not a tech nerd like you so please feel free to be granular with your explanation here)
2. Is the database composed of only human studies or both mice and human studies? If both, is there a way we give more priority to data from human studies?
3. Can you explain a little further how recommendations are generated? Are recommendations EXPLICITLY based on peer reviewed studies?
4. How often is new data collected from patients/customers used to update the database? (In this case for MP)
5. The algorithm/tech (how proprietary it is, how it learns/trains, who developed it, etc)
Strongly Suggested Readings
The following posts and video are strongly recommended before reading the answers below. Why? People often come into this are with simplistic naïve beliefs. These often end up with “Why don’t you have… ” The answer is usually, there is not the data or the data we have is very nuisance and cannot be safely applied outside of the pre-conditions for the study.
- Mankind does not have an ideal Microbiome
- The shortfall in available probiotics
- The taxonomy nightmare before Christmas…
To test your understanding of the above try answering the following questions (answers at bottom of the page)
- A: Two studies of depression were done in India, should the results be the same? I.e. confirming results?
- B: A probiotic was developed for depression and tried in India and Iceland. One had strong statistical significance and the other did not despite both being large studies. Is this likely to occur?
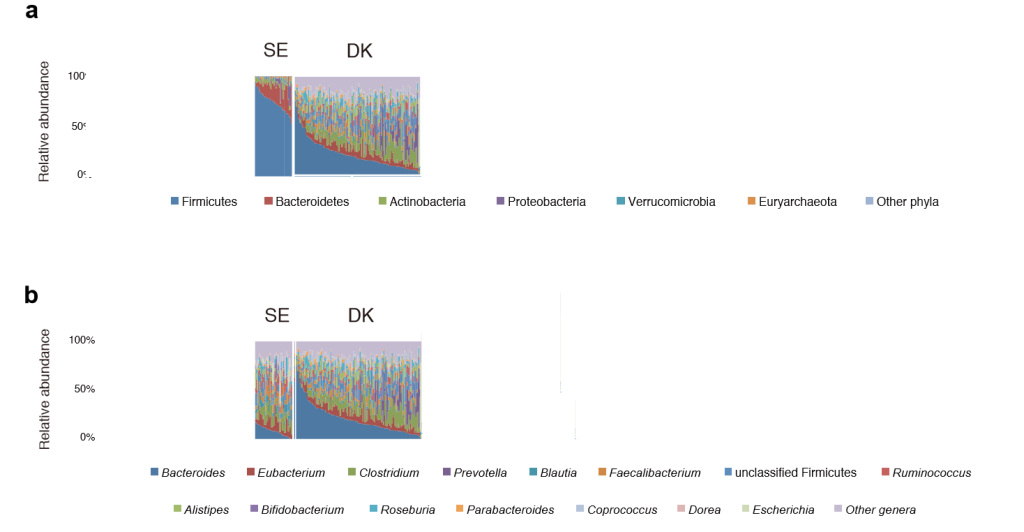
- C: Two studies are done on depression, one in Denmark(DK) and one in Sweden (SE), both study used the same lab in Germany. Is it likely they will produce the same results? “After all the people have the same DNA and eat the same things”
Answer to Question #1
How does the algorithm/tech learn and train? (I am a biologist and not a tech nerd like you so please feel free to be granular with your explanation here)
There is no training. Training applies to a branch of Artificial Intelligence called Machine Learning. Machine learning requires large well scoped dataset — for example, people with depression in South Africa that are Seventh Day Adventists with less than 25% Caucasian inheritance. “At a bare minimum, collect around 1000 examples. · For most “average” problems, you should have 10,000 – 100,000 examples. “[ How much data is required for machine learning?]
The nature of this problem space is not suitable for machine learning. It is suitable for a different branch of Artificial Intelligence. This branch applies a Mixed Integer Programming solution(MIP) that uses fuzzy logic(FL) integrated with Monte Carlo simulation (MC) to handle issues with bacteria selection, significance of each bacteria (include or exclude from the MIP), and the weight of each bacteria (parameters of the MIP). There are many many ways to select bacteria.
- The classic way is to used existing published human studies — but those results are sensitive to experiment construction (i.e. populations) as well as which of 97+ different reports you can get according to which lab, lab software and data version is used.
- We include this information into the Monte Carlo simulation when appropriate and available
- The alternative that we use is doing apples-to-apples, i.e. using data that has been processed with the same lab, lab software and data version. We call these Special Studies. Some of the assumptions being used can be nit-picked extensively, but it is the best data that’s available for microbiome data obtained easily and at a reasonable expense.
Answer to Question #2
Is the database composed of only human studies or both mice and human studies? If both, is there a way we give more priority to data from human studies?
The amount of data available from human studies is actually very sparse. Many studies come from veterinary practice (not just mice alone, dogs, cats, sheep, cattle, etc). Early, we encountered some substances that had opposite effects on different human populations. The best contrast are the many studies coming out of China versus Studies out of the US or the UK — different eating habits and DNA come into play.
Additionally, if the criteria is human studies, then gender and age are significant factors for how a substance changes. Many studies fail to declare that information or provide data broken down by these factors.
An early decision evaluating the volume of data from various criteria resulted in the inclusion criteria being a simple “any mammal study” and for a few very special cases, lab studies using reactors only. If Barley causes a certain bacteria to increase in mice, it is likely that it will cause the same in human. Note the use of the word “likely”. For some substances we may have over two dozen studies, we use these studies to compute a fuzzy logic coefficient on the confidence that it will shift.
Answer to Question #3
Can you explain a little further how recommendations are generated? Are recommendations EXPLICITLY based on peer reviewed studies?
Depending on the user (or a wholesale customer), this may be done in various ways to select the inclusion criteria.
- The simplest case is human studies published on the US National Library of Medicine (PUBMED).
- Most of these studies report only which bacteria (at various taxonomy levels) were found to be statistically significant above or below the average of the control population.
- This does not provide clarity on whether being 1 or 10 Standard Deviation above the control population is significant for an individual sample. The study used the means of a group compared to the means of another group.
- A variety of criteria could be applied to determine if a level is significant for a specific sample for a person:
- Actual Percentile Ranking
- Z-Score Ranking
- Box-Plot Whiskers
- and many more
- Another way is to use “reference ranges” from some authority, for example, Dr. Jason Hawrelak or ranges provided from the lab or other sources. This simplifies the above issue of what is significant, but not all bacteria list in PUBMED may have reference ranges. Additionally, the ranges may be dependent of a specific lab being used, for example, Dr. Jason Hawrelak ranges were developed using results from uBiome. These reference ranges may be used isolated from PUBMED findings because they are deemed to be for “a healthy microbiome” based on the population they were developed for.
There are many many other paths possible. Some of the processes we used are proprietary and derived from economics and finance.
So the answer is: YES, they can be based solely on peer review studies. Our experience has not been good using that criteria, i.e. cross validation on the suggestions were a poor fit against clinical trials of substances. Further more, the study, Altered fecal microbiota composition in patients with major depressive disorder. Brain, behavior, and immunity (Brain Behav Immun ) Vol: 48 Issue Pages: 186-94, on depression finds that Francisella tularensis (NCBI:263 ) is low. Finding substances from human peer review studies for substances that impacts this bacteria failed to find any studies that will raise this bacteria. Extrapolation from existing studies on PubMed is required.
Answer to Question #4
How often is new data collected from patients/customers used to update the database? (In this case for MP)
Information from PUBMED is updated once a week. Statistics (specific for each lab) is recomputed once a week as are results from Special Studies.
Answer to Question #5
The algorithm/tech (how proprietary it is, how it learns/trains, who developed it, etc)
The algorithms were developed by Kenneth Lassesen, M.Sc. over the last 5 years of repeated tuning by doing cross validation of suggestions for some conditions against clinical studies of the same suggestions being used on those conditions. The implementation is a strict trade secret. The algorithms are continuously being updated based on new studies (public and confidential ones). The general approach is described in Question #1.
Answers to Reading Comprehension Questions Above
A: It is very unlikely, one study could be done on strict vegan Hindu who are urban dwellers, the other study could be done Muslin rural workers near the sea (thus meat and fish in their diet). Additionally each could be using different labs. Re-read The taxonomy nightmare before Christmas…
B: It is very likely, see A: above. Additionally, how the probiotic was sourced from humans can be very significant. Re-read The shortfall in available probiotics
C: In this case, unlikely, because the microbiome of these apparently similar population are different as illustrated below. Re-read Mankind does not have an ideal Microbiome
Recent Comments