Using my last post, The root of all evil in the Microbiome?, I tried identifying items that address many of the evils. The process is simple, I use Microbiome Prescription existing database with 2,099.126 interactions between gut modifiers and bacteria. I then select items that only increase the top N items. Starting with 2 items and then working up.
Two Items: Prevotella, Veillonella
Produced following items
- Amino Acid and similar
- acetic acid
- l-citrulline
- propionate
- Antibiotics, Antivirals etc
- rifaximin (antibiotic)s
- Diet Style
- ketogenic diet
- Food (excluding seasonings)
- blackcurrant
- cranberry bean flour
- Far infrared Sauna
- fasting
- lupin seeds (anaphylaxis risk, toxic if not prepared properly)
- Miso
- pea (fiber, protein)
- Pulses
- red wine
- wheat bran
- Herb or Spice
- Bofutsushosan
- chinensis (Chinese goldthread)
- isobutyric acid
- isovaleric acid(fatty acid)
- smoking
- Prebiotics and similar
- arabinoxylan oligosaccharides (prebiotic)
- galacto-oligosaccharides (prebiotic)
- xylan (prebiotic)
- Probiotics
- Lentilactobacillus buchneri
- Sugar and similar
- non-starch polysaccharides
- saccharin
- Vitamins, Minerals and similar
- vitamin d
Three Items: Prevotella, Veillonella, Clostridium
We are done to just one item: Amino Acid and similar l-citrulline
This is not unexpected, in building the suggestions algorithms, I often encountering items that helps one and hurts another; also a lack of studies.
Alternative Approach
I am going to pick the top dozen (12) genus, and then select the items by the number of genus positively impacted. As you see, the best that we get is impacting 50% of the genus picked.
Remember: we are doing items with absolutely positive impact only. We exclude all items that has a negative impact on any of the genus reported in any study. For clarity, if a substance increases a genus in some studies and decreases in other studies, it is excluded. This is the most conservative approach.
Caution: this assumes undergrowth for all 12 genus. It may make an overgrowth worse.
| ModifierType | Modifier2 | Cnt |
| Prescription – Other | proton-pump inhibitors (prescription) | 6 |
| Sugar and similar | non-starch polysaccharides | 5 |
| Amino Acid and similar | acetic acid | 3 |
| Amino Acid and similar | Conjugated Linoleic Acid | 3 |
| Amino Acid and similar | l-citrulline | 3 |
| Amino Acid and similar | proline (amino acid) | 3 |
| Diet Style | animal-based diet | 3 |
| Food (excluding seasonings) | blackcurrant | 3 |
| Food (excluding seasonings) | fasting | 3 |
| Food (excluding seasonings) | lupin seeds (anaphylaxis risk, toxic if not prepared properly) | 3 |
| Food (excluding seasonings) | Pulses | 3 |
| Food (excluding seasonings) | safflower oil | 3 |
| Food (excluding seasonings) | wheat bran | 3 |
| Miscellaneous, food additives, and other odd items | isobutyric acid | 3 |
| Miscellaneous, food additives, and other odd items | isovaleric acid(fatty acid) | 3 |
| Prebiotics and similar | arabinoxylan oligosaccharides (prebiotic) | 3 |
| Sugar and similar | saccharin | 3 |
| Amino Acid and similar | propionate | 2 |
| Diet Style | fibre-rich macrobiotic ma-pi 2 diet | 2 |
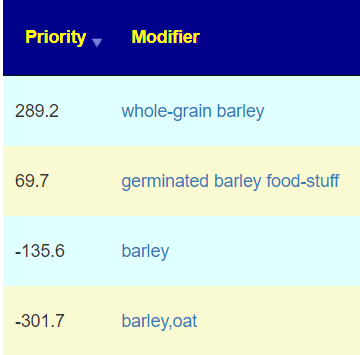
| Food (excluding seasonings) | barley,oat | 2 |
| Food (excluding seasonings) | cranberry bean flour | 2 |
| Food (excluding seasonings) | Miso | 2 |
| Food (excluding seasonings) | navy bean | 2 |
| Food (excluding seasonings) | pectin | 2 |
| Food (excluding seasonings) | red alga Laurencia tristicha | 2 |
| Herb or Spice | Bofutsushosan | 2 |
| Herb or Spice | coptis chinensis (Chinese goldthread) | 2 |
| Herb or Spice | plantago asiatica l. | 2 |
| Herb or Spice | schisandra chinensis(magnolia berry or five-flavor-fruit) | 2 |
| Miscellaneous, food additives, and other odd items | Tributyrin | 2 |
| Prebiotics and similar | carboxymethyl cellulose (prebiotic) | 2 |
| Prebiotics and similar | oligofructose (prebiotic) | 2 |
| Prebiotics and similar | resistant maltodextrin | 2 |
| Prescription – Other | epicor | 2 |
| Probiotics | bacillus subtilis natto (probiotics) | 2 |
| Probiotics | saccharomyces boulardii (probiotics) | 2 |
| Sugar and similar | glucose (sugar) | 2 |
| Sugar and similar | levan | 2 |
| Vitamins, Minerals and similar | Ferric citrate | 2 |
Bottom Line
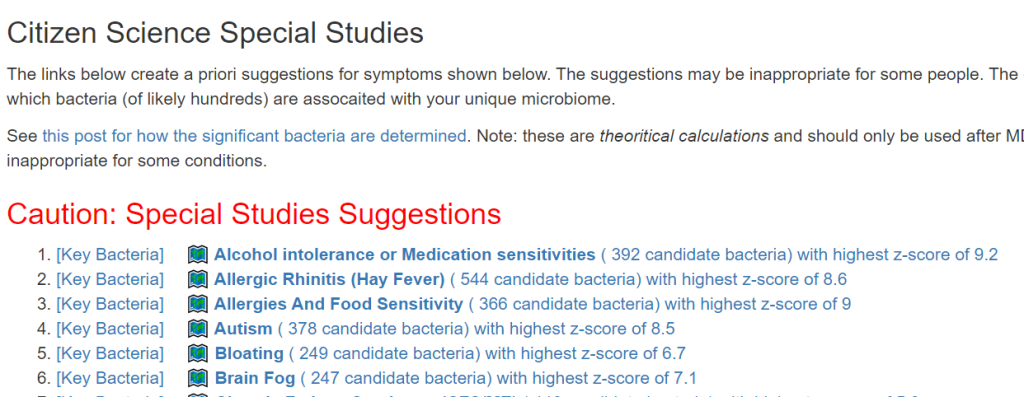
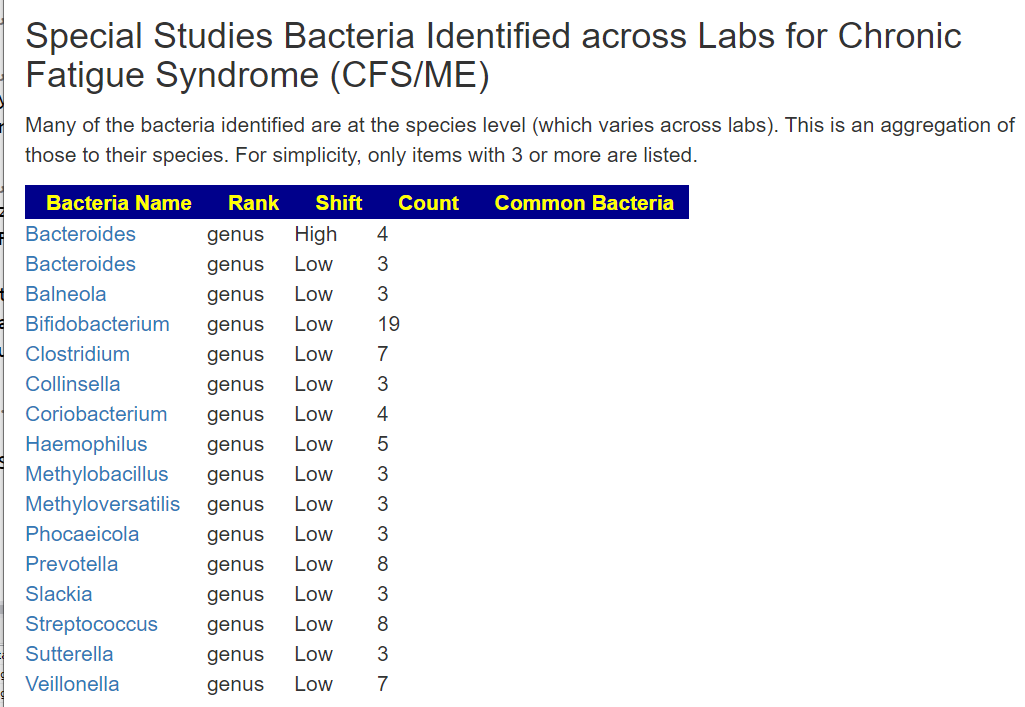
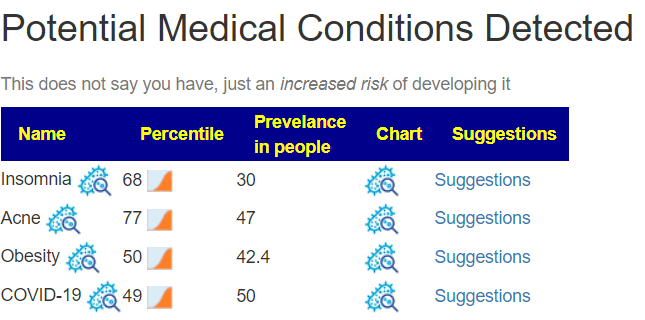
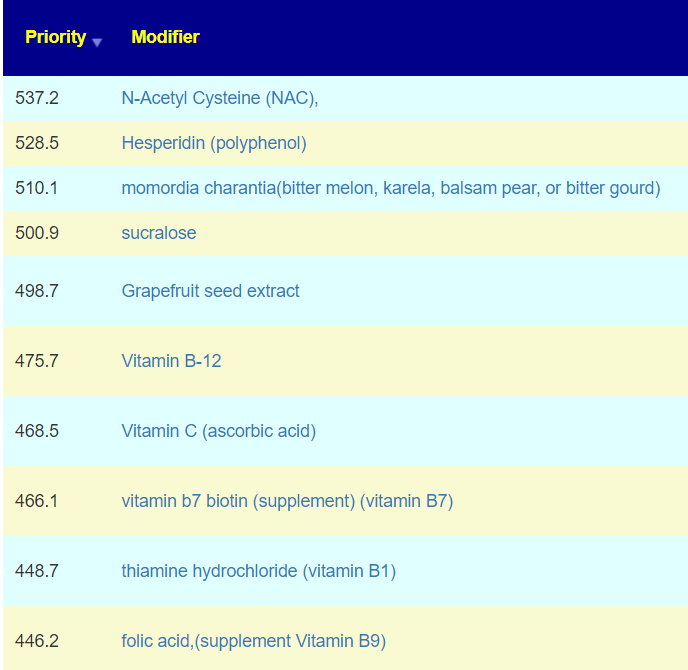
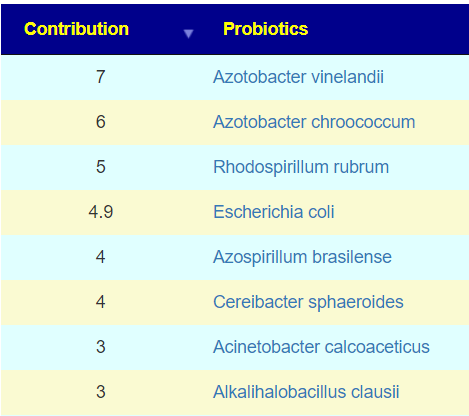
The above is a generic list that should “do no harm” for most people and will likely help. Better results are likely to occur using a 16s microbiome report (OmbreLabs [US only] and Biomesight [World wide] are the most used) and using the Artificial Intelligence on Microbiome Prescription that factors in the 2 million facts that it has available (see this example). That would produce a targeted list for your needs.
Recent Comments