As a result of doing an analysis for a 19 month old toddler, I added a new option that can also be used with Transcribed tests. This post applied to the following tests:
- Bioscreen
- Biovis Microbiome Plus
- DayTwo
- Diagnostic Solution GI-Map
- GanzImmun Diagnostic A6
- GanzImmun Diagnostics AG Befundbericht
- Genova Gi Effects
- Genova Parasitology
- GI EcologiX (Invivo)
- GI360 Stool (UK)
- Gut Zoomer
- InVitaLab
- Kyber Kompakt
- Medivere Mikrobiom Plus Stuhlanalyse
- Medivere: Darm Mikrobiom Stuhltest
- Medivere: Darn Magen Diagnostik
- Medivere: Gesundsheitscheck Darm
- Metagenomics Stool (De Meirleir)
- Nordic Laboratories
- NutriPATH
- Smart Gut
- Verisana
You must save your input.
Process
Here’s an example
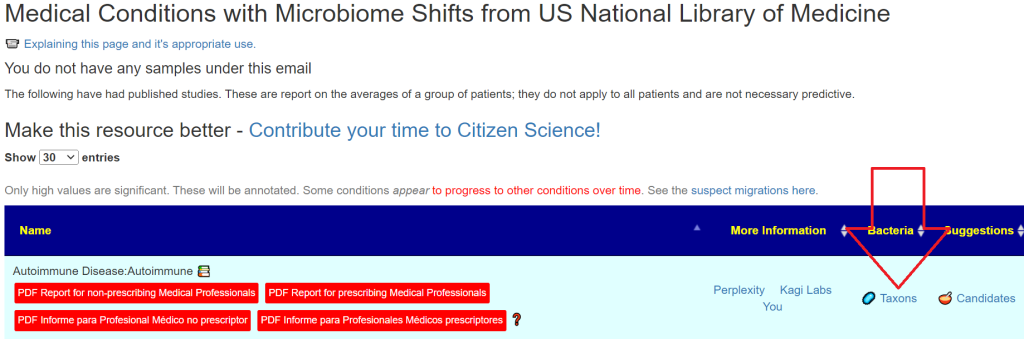
When you logged in, you will see your saved tests, CLICK ON Review.
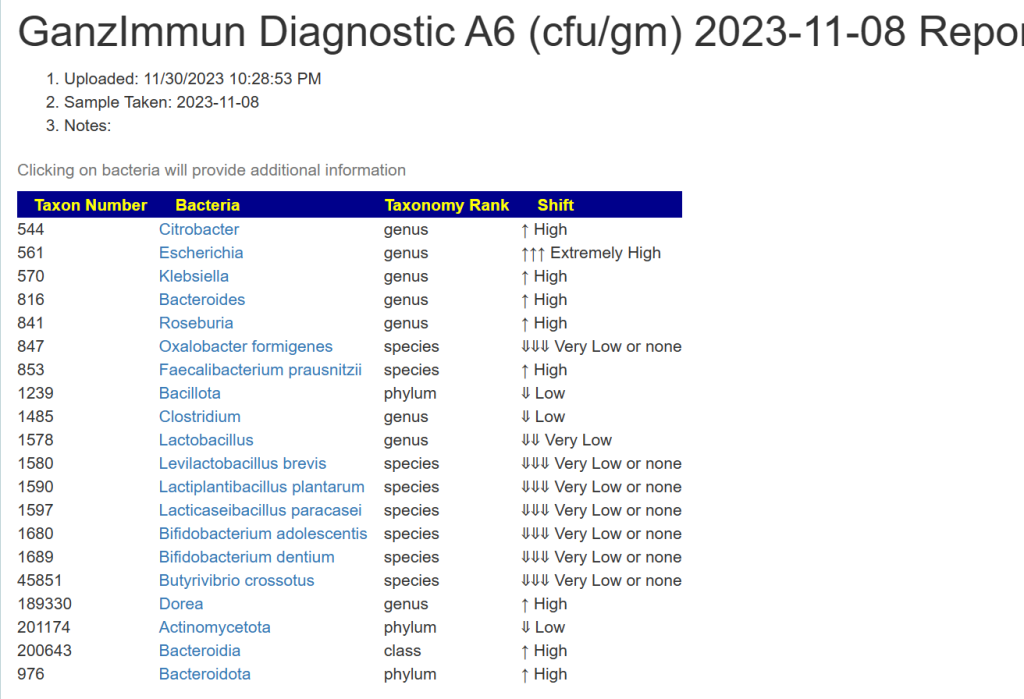
And then we have the details you entered below with an important column, taxon number.
Below this are conditions where your pattern matches at least 5 shifts reported in Published Studies.
There may be many items listed. This is by pattern matching and is not predictive.
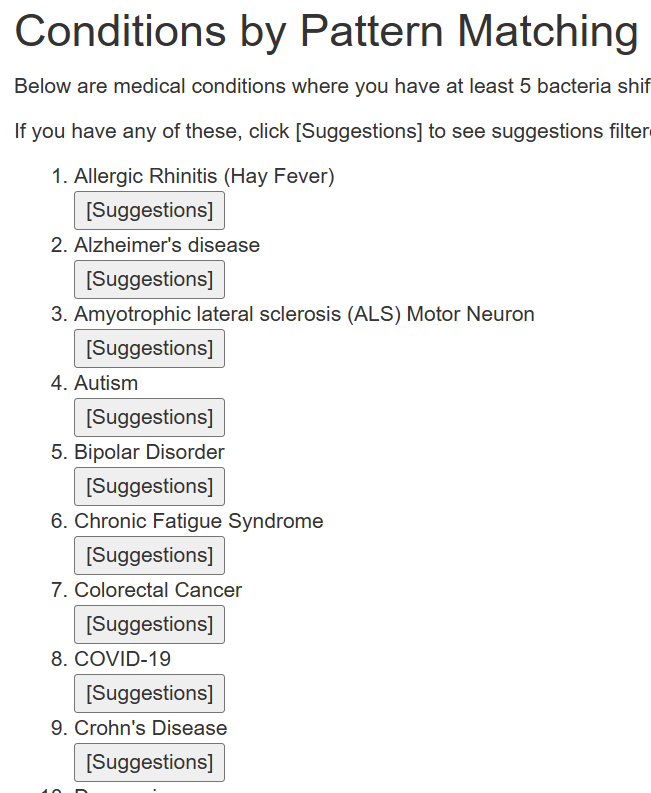
If you have any of these conditions, or suspect you may have. Just click the appropriate button.
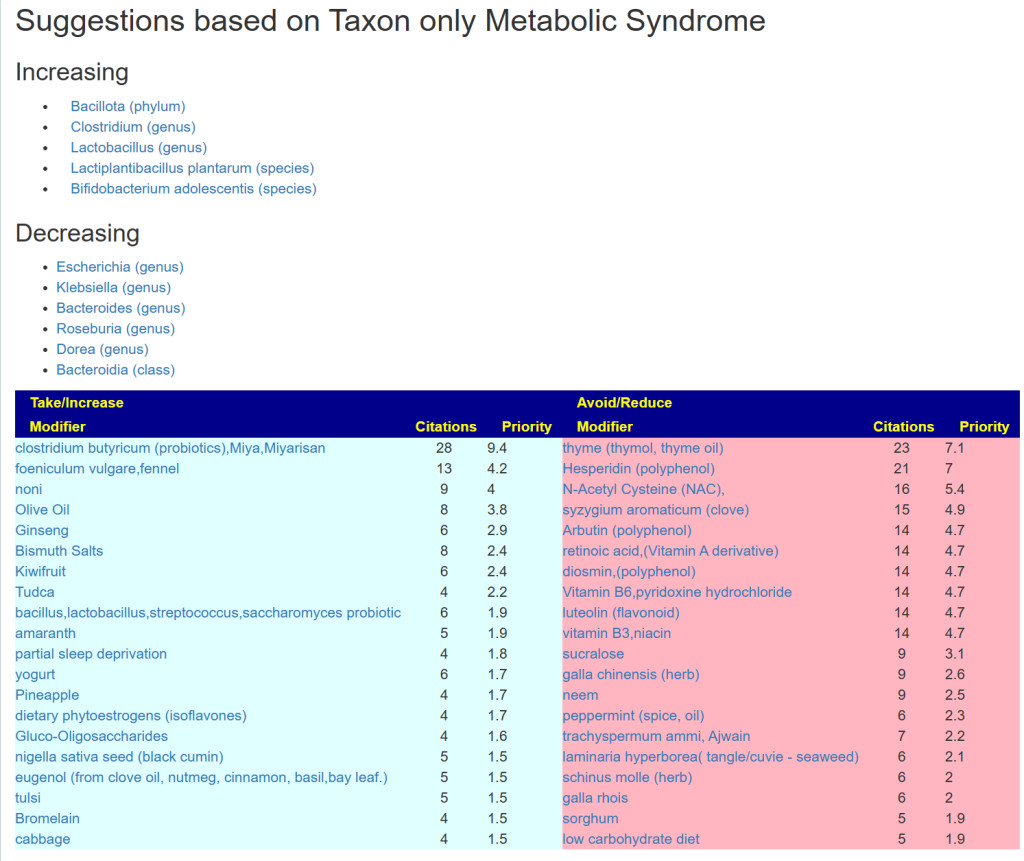
An example is below. These are tuned safest-suggestions for the matches. What do I mean by safest? It means the items are not reported in any study in the database to adversely impact any of bacteria listed. Many substances have contradictory reports on shifts — this substances are excluded.
Not Listed Condition?
This person believes they may have Autoimmune, so going to https://microbiomeprescription.com/Library/PubMed we find that it is listed.
If it is not listed, search for bacteria shifts reported and use those (please send me the studies so I may add them).
The bacteria are shown in a tree. You have to manually match between the two.

In this we have:
- Escherichia ⬇️ but our sample is high,
- Roseburia intestinalis ⬆️ - we are high on Roseburia, we will include it
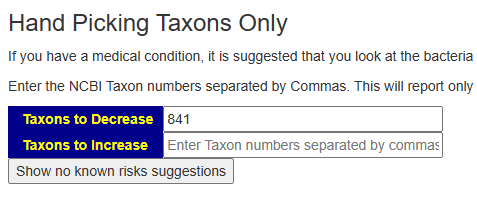
We have only one match — this tests with limited reporting is not a good fit for this condition. Doing a test like Biomesight, Xenogene, Thorne or Ombre is likely the best choice.
We just copy the taxon number into the form at the bottom of the page, and then click suggestions.
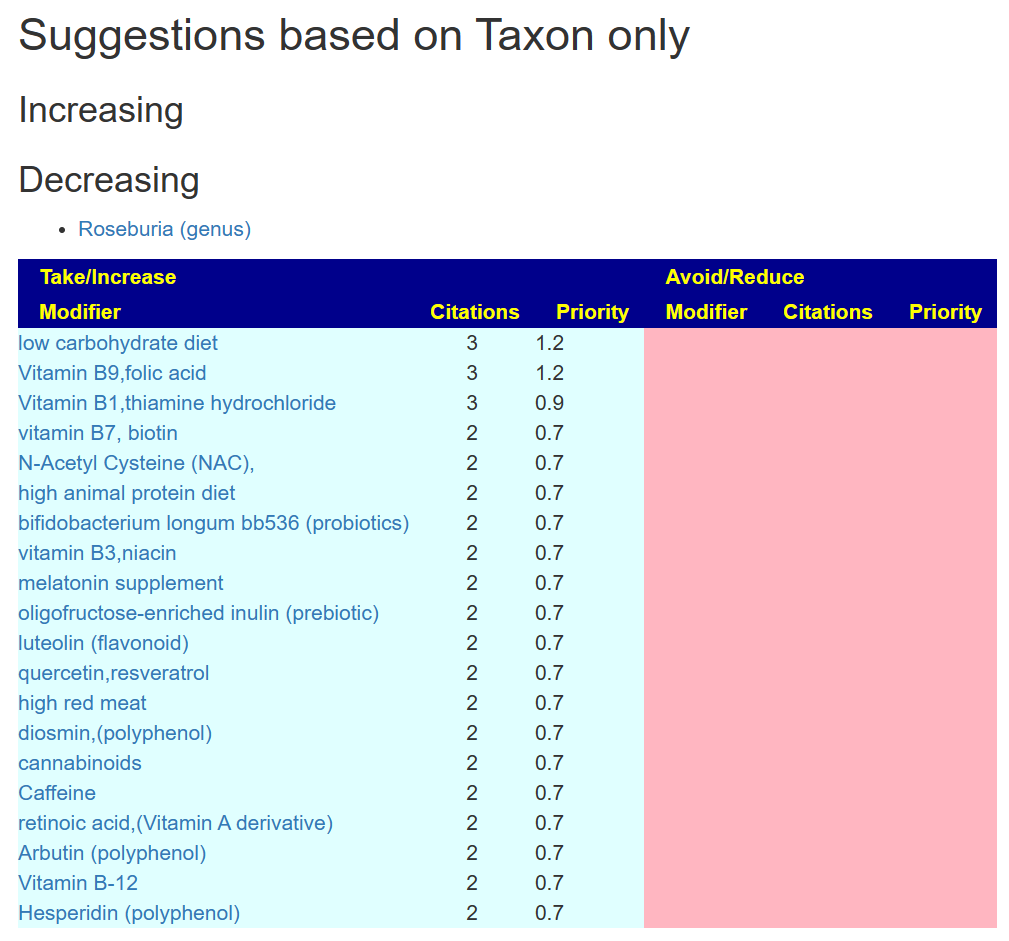
In this case, we get a short list. Remember, doing a single bacteria means you are ignoring a lot of interactions and factors. The suggestions could feed other bacteria that are too high.
Recent Comments