Recently I did a much more rigorous implementation of the Kaltoft-Moldrup ranges as part of a periodic review and refactor. Today, I tested a hypothesis that those ranges may be good for detecting the taxa (bacteria) associated with a self-reporting symptom. The results exceeded expectations.
The methodology is simple to understand. Suppose we have 1000 samples. The KM lower limit is 5%ile and upper limit is 85%ile across the entire population.
For those with brain fog as a symptom we found that instead of the expected 5% of 1000 (i.e. 50), we only have just 5 samples in this range. That is a dramatic (and statistically significant shift).
On the other end, we do not have our expected 150 (100%-85% * 1000) but 300 sample in this range.
We can conclude that those with less than the 5%ile is associated with brain fog as well as those with more than 85%ile.
Most of the time we will find just one shift being significant. At times, both can be — i.e. values cascaded away from the range is both directions.
I have determined the taxa/bacteria for a stack of symptoms using P < 0.01. Because different labs identifies taxa information (see Nightmare ). This is why there is often no replication of (or contrary) results in many published studies. I created a table that is lab specific for each taxa.
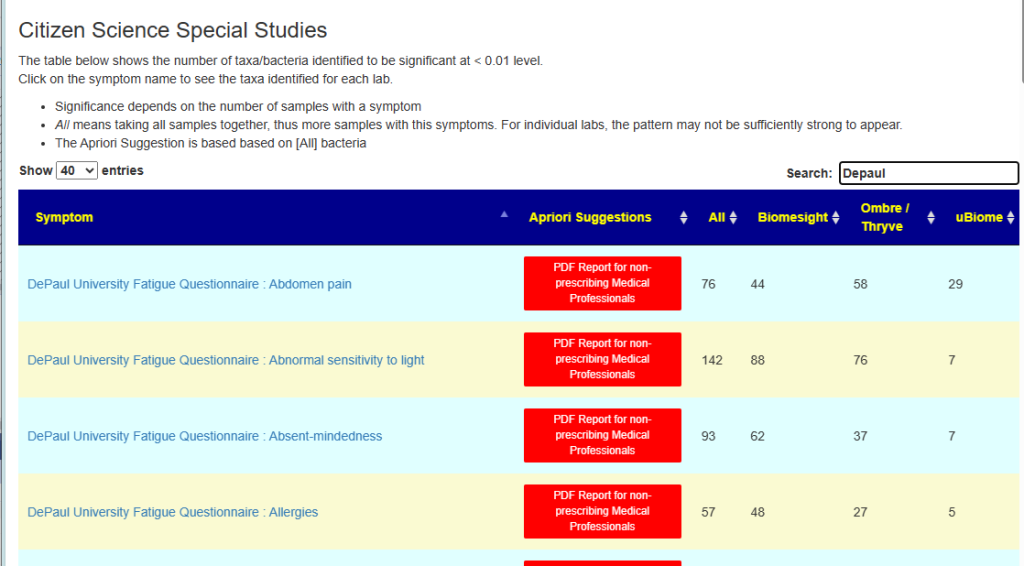
Overview Page
This shows the number of bacteria found significant for each symptom. “All” usually will have more because it has more samples to use and thus can detect lesser associations better.
Link: https://microbiomeprescription.com/Library/CitizenScience
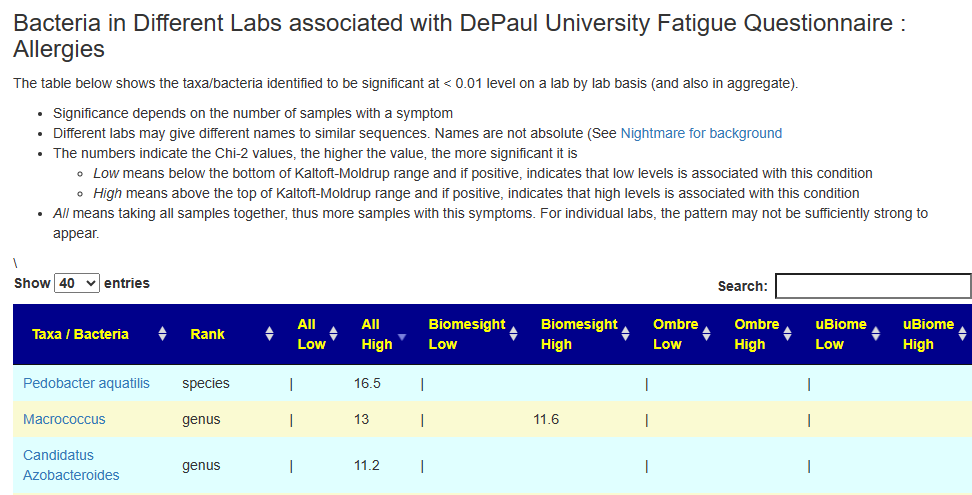
Detail Page
This page can get confusing because the labs use different sequences of RNA to infer the bacteria. The actual association
PDF A Priori Suggestions
This is done using the ALL profile.
2 thoughts on “New Special Studies on Symptoms”
Comments are closed.