Time to Refresh What we know
A reader request with severe MCS motivated me to revisit Multiple Chemical Sensitivity (MCS) and literature on it. I am first going to recap my memory of what I know, a literature review, Last, analysis using data contributed to Microbiome Prescription – which did not find anything major beyond bacteria shifts that are statistically significant. We explore enzymes, products and substrates with no smoking guns found
There was a processing error — This post was revised
Long weekend tracing issue, made short. Some revisions to reduce running cost had a typo pointing one set of data (Substrate data for Samples) to the wrong lookup table to determine percentile. This has been corrected and the data audited to insure correctness.
I had mild MCS during one flare. It disappeared with remission.
Multiple Chemical Sensitivity (MCS) is not bronchial (breathing). There was a series of studies where the person was wearing a half-face mask breathing compressed air. Bringing a triggered close to the person (blind testing), trigger the response. My own wife lab results leaves a significant scent. She had a complete workup with Dave Berg, Hemex Lab and two weeks later she had a bad MCS reaction. About a week later, new blood work was sent. Everything was the same, except for a marker indicating very active coagulation. The marker had a short half-life around 2 hours. After she fully recovered, same test returned to normal. This hints at coagulation being part of the process.
– Ken Lassesen
We did find one effective treatment for shortening the duration of a MCS episode: Olestra (as in Olestra chips). This compound sucks chemicals out of tissues [2015] [2014]
I attended a talk today with the AAAS (I’m a member) and Memory Cells was discuss for some autoimmune conditions. It occurs to me that these are likely a factor. These cells become uber-sensitive to specific chemical compounds and have a lasting memory. In time they may die off, but not quickly.
Caution: this post is subject to revision. The findings to date are so significant that I am posting early.
The Current Literature
First a summary from a 2022 review – it covers a lot of territory… many quotes are below
Multiple Chemical Sensitivity (MCS), a condition also known as Chemical Sensitivity (CS), Chemical Intolerance (CI), Idiopathic Environmental Illness (IEI) and Toxicant Induced Loss of Tolerance (TILT), is an acquired multifactorial syndrome characterized by a recurrent set of debilitating symptoms. The symptoms of this controversial disorder are reported to be induced by environmental chemicals at doses far below those usually harmful to most persons. They involve a large spectrum of organ systems and typically disappear when the environmental chemicals are removed. However, no clear link has emerged among self-reported MCS symptoms and widely accepted objective measures of physiological dysfunction, and no clear dose-response relationship between exposure and symptom reactions has been observed. In addition, the underlying etiology and pathogenic processes of the disorder remain unknown and disputed, although biologic and psychologic hypotheses abound. It is currently debated whether MCS should be considered a clinical entity at all.
Multiple Chemical Sensitivity [2022]
- “Not surprising, MCS is often juxtaposed to, and sometimes combined with, other diseases for which definitive diagnoses and explanations are also found wanting, including fibromyalgia (FM), Gulf War syndrome (GWS), chronic fatigue syndrome (CFS), sick building syndrome (SBS), and electromagnetic radiation exposure (ERE) [18,23,24,25,26,27,28,29,30,31,32].”
- “An association was documented, however, between MCS and increased nasal airflow resistance, respiration rate, and heart rate.” [2021] Volatile chemicals can not only affect respiration, but can cause laryngeal symptoms which, in the extreme, can induce vocal cord dysfunction [82].
- Changes in skin conductance were also seen in MCS patients but not in controls [193].
- Compared to younger persons, those 65 years of age and older are less likely to identify themselves as chemically sensitive [88].
- In accord with this concept, McKeown-Eyssen et al. [219] found, in a female cohort of 203 MCS cases and 162 controls, that the cases had higher levels of cytochrome P450 CYP2D6 (i.e., one of the most important enzymes involved in the metabolism of xenobiotics in the body) and n-acetyl transferase 2 (NAT2). More possible genes listed here.
- Like the sick building syndrome [89], there is a remarkably higher prevalence in women than in men [16,18,21,90,91], with the percentage of women ranging from 60% to 88% [18,57,92,93,94].

- More than half (59%) of the mast cell activation syndrome group met criteria for chemical intolerance. [2021]
- Moist and Mold Exposure is Associated With High Prevalence of Neurological Symptoms and MCS in a Finnish Hospital Workers Cohort [2020]
Microbiome and MCS
Almost every condition in the above chart has microbiome shifts associated with it. Exposure to mold also is known to cause microbiome shifts. There appears to be no published studies exploring this possibility. At this time we have 128 people (7.3%) who have annotated their samples with Multiple Chemical Sensitivity [uBiome: 55, Ombre:39, Biomesight 25] to Microbiome Prescription.A
Executing analysis by lab, we found only one taxon reported significant in two labs. Bacteroides clarus where people with MCS had 3x the count of people not reporting MCS [BiomeSight, uBiome]. The top items of significance are shown below. All of them have much higher counts in people with MCS.
| Source | probability | Tax_Name | tax_rank | with MCS | without MCS | Obs |
| BiomeSight | P < 0.001 | Anaerotruncus | genus | 3356 | 1834 | 683 |
| BiomeSight | P < 0.001 | Anaerotruncus colihominis | species | 3208 | 1743 | 683 |
| BiomeSight | P < 0.001 | Bacteroides intestinalis | species | 28980 | 2430 | 396 |
| Thryve | P < 0.001 | Prevotella maculosa | species | 27399 | 5733 | 404 |
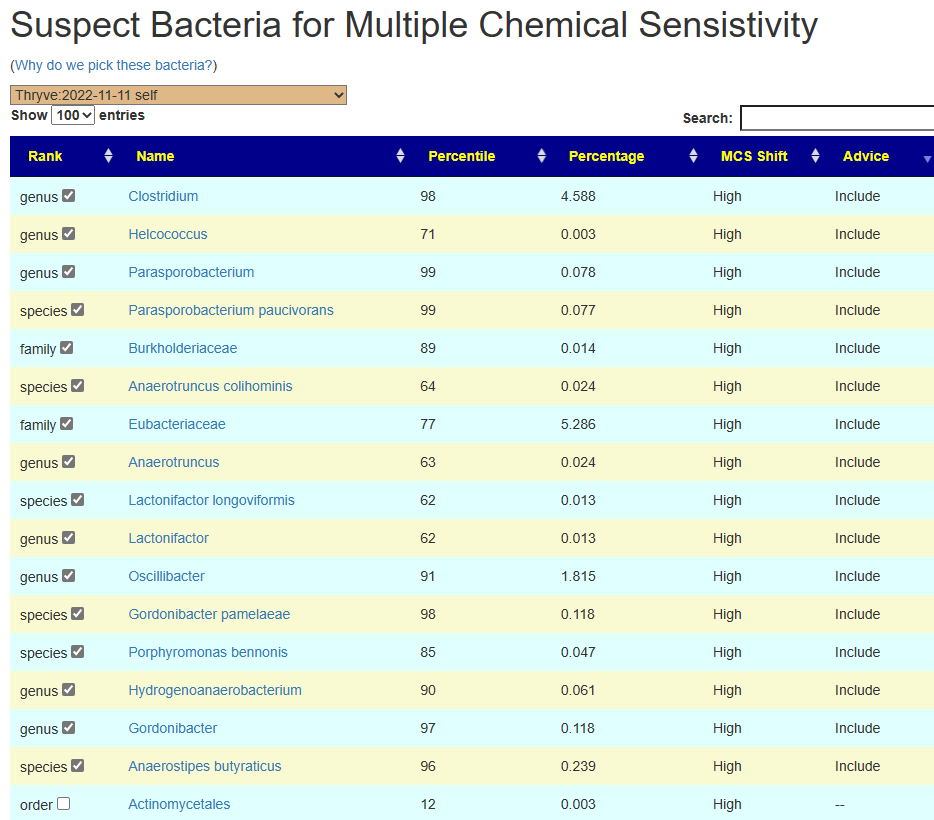
These have been added to the [Research] tab, as shown below.
Looking at KEGG Enzymes we have the strongest candidates below. Looking for any enzyme that is significant for 2 or more labs, we had only one: 5.3.1.15 D-lyxose aldose-ketose-isomerase (Biomesight and Thryve).
| EnzymeName | ECKEY | Src | WithMean | WithoutMean | TScore | DF |
| 7alpha-hydroxysteroid:NAD+ 7-oxidoreductase | 1.1.1.159 | BiomeSight | 14681 | 2479 | 4.01 | 657 |
| acetyl-CoA:glycerone phosphate C-acetyltransferase | 2.3.1.245 | BiomeSight | 49551 | 27444 | 3.56 | 701 |
| acetyl-CoA:heparan-alpha-D-glucosaminide N-acetyltransferase | 2.3.1.78 | BiomeSight | 26909 | 9394 | 4.02 | 671 |
| alginate beta-D-mannuronate—uronate lyase | 4.2.2.3 | BiomeSight | 26933 | 9472 | 4.02 | 681 |
| CTP:phosphoglutamine cytidylyltransferase | 2.7.7.103 | BiomeSight | 27471 | 13156 | 3.97 | 702 |
| L-alanine:glyoxylate aminotransferase | 2.6.1.44 | uBiome | 16083 | 2029 | 3.40 | 309 |
| NAD+:glycine ADP-D-ribosyltransferase (sulfide-adding) | 2.4.2.59 | BiomeSight | 48398 | 26181 | 3.60 | 699 |
| phosphoenolpyruvate:glycerone phosphotransferase | 2.7.1.121 | BiomeSight | 68289 | 41808 | 3.60 | 702 |
| sn-glycerol-1-phosphate:NAD(P)+ 2-oxidoreductase | 1.1.1.261 | BiomeSight | 29070 | 11440 | 3.55 | 700 |
Dropping by Lab and using Percentiles Only
Trying a different approach aggregating all of the data and using percentile ranking. This masks most of the differences between labs and gives us large sample size, We have a very actionable item: increase Bifidobacterium by taking Bifidobacterium probiotics.
| tax_name | tax_rank | percentile | Obs |
| Alistipes indistinctus | species | 61.7 | 61 |
| Anaerobutyricum hallii | species | 39.8 | 41 |
| Bifidobacteriales | order | 37.7 | 106 |
| Bifidobacterium | genus | 37.1 | 122 |
| Oscillospiraceae incertae sedis | norank | 63.1 | 41 |
| Acidobacteria | phylum | 33.2 | 48 |
| Butyricimonas faecihominis | species | 61.2 | 43 |
| Bifidobacteriaceae | family | 37.4 | 122 |
| Collinsella aerofaciens | species | 39.1 | 83 |
| Phascolarctobacterium succinatutens | species | 37.3 | 52 |
| Hungateiclostridiaceae | family | 61.4 | 45 |
KEGG Enzymes
Applying this to KEGG Enzymes, we have just one of concern that is high: EC3.2.1.46 galactosylceramidase, and a big 39 that are low indicating a deficiency of enzymes is likely
| EnzymeName | percentile | obs |
| [2.7.1.85] ATP:cellobiose 6-phosphotransferase | 34.6 | 64 |
| [4.1.1.31] phosphate:oxaloacetate carboxy-lyase (adding phosphate, phosphoenolpyruvate-forming) | 35.2 | 122 |
| [3.2.1.199] 6-sulfo-alpha-D-quinovosyl diacylglycerol 6-sulfo-D-quinovohydrolase | 36.1 | 61 |
| [1.17.2.1] nicotinate:cytochrome 6-oxidoreductase (hydroxylating) | 36.7 | 63 |
| [3.5.1.18] N-succinyl-LL-2,6-diaminoheptanedioate amidohydrolase | 37.9 | 122 |
| [2.7.1.211] protein-Npi-phospho-L-histidine:sucrose Npi-phosphotransferase | 38 | 122 |
| [1.2.1.99] 4-(gamma-L-glutamylamino)butanal:NAD(P)+ oxidoreductase | 38.1 | 63 |
| [1.17.98.4] formate:[oxidized hydrogenase] oxidoreductase | 38.3 | 121 |
| [3.4.23.51] | 38.4 | 117 |
| [2.7.1.195] protein-Npi-phospho-L-histidine:2-O-alpha-mannopyranosyl-D-glycerate Npi-phosphotransferase | 38.4 | 94 |
| [1.1.1.65] pyridoxine:NADP+ 4-oxidoreductase | 38.4 | 98 |
| [2.1.1.172] S-adenosyl-L-methionine:16S rRNA (guanine1207-N2)-methyltransferase | 38.5 | 120 |
| [2.1.1.264] S-adenosyl-L-methionine:23S rRNA (guanine2069-N7)-methyltransferase | 38.6 | 121 |
| [3.5.1.32] N-benzoylamino-acid amidohydrolase | 38.7 | 63 |
| [2.7.7.12] UDP-alpha-D-glucose:alpha-D-galactose-1-phosphate uridylyltransferase | 38.7 | 122 |
| [2.4.1.342] ADP-alpha-D-glucose:alpha-D-glucose-1-phosphate 4-alpha-D-glucosyltransferase (configuration-retaining) | 38.9 | 121 |
| [4.1.2.22] D-fructose-6-phosphate D-erythrose-4-phosphate-lyase (adding phosphate; acetyl-phosphate-forming) | 39 | 122 |
| [4.1.2.9] D-xylulose-5-phosphate D-glyceraldehyde-3-phosphate-lyase (adding phosphate; acetyl-phosphate-forming) | 39 | 122 |
| [3.2.1.122] alpha-maltose-6′-phosphate 6-phosphoglucohydrolase | 39 | 122 |
| [1.1.1.9] xylitol:NAD+ 2-oxidoreductase (D-xylulose-forming) | 39.1 | 83 |
| [6.3.1.12] D-aspartate:[beta-GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)]n ligase (ADP-forming) | 39.2 | 122 |
| [4.1.2.40] D-tagatose 1,6-bisphosphate D-glyceraldehyde-3-phosphate-lyase (glycerone-phosphate-forming) | 39.2 | 122 |
| [3.7.1.12] cobalt-precorrin 5A acylhydrolase | 39.2 | 122 |
| [1.3.1.28] (2S,3S)-2,3-dihydro-2,3-dihydroxybenzoate:NAD+ oxidoreductase | 39.2 | 73 |
| [5.3.1.26] D-galactose-6-phosphate aldose-ketose-isomerase | 39.3 | 115 |
| [2.7.1.19] ATP:D-ribulose-5-phosphate 1-phosphotransferase | 39.4 | 62 |
| [2.7.1.208] protein-Npi-phospho-L-histidine:maltose Npi-phosphotransferase | 39.4 | 122 |
| [2.7.1.207] protein-Npi-phospho-L-histidine:lactose Npi-phosphotransferase | 39.4 | 118 |
| [3.2.1.185] beta-L-arabinofuranoside non-reducing end beta-L-arabinofuranosidase | 39.4 | 72 |
| [1.1.1.130] 3-dehydro-L-gulonate:NAD(P)+ 2-oxidoreductase | 39.5 | 72 |
| [3.2.1.85] 6-phospho-beta-D-galactoside 6-phosphogalactohydrolase | 39.5 | 117 |
| [5.1.1.10] amino-acid racemase | 39.7 | 108 |
| [3.2.1.10] oligosaccharide 6-alpha-glucohydrolase | 39.7 | 122 |
| [1.1.1.203] uronate:NAD+ 1-oxidoreductase | 39.7 | 61 |
| [3.5.2.9] 5-oxo-L-proline amidohydrolase (ATP-hydrolysing) | 39.7 | 120 |
| [2.7.1.191] protein-Npi-phospho-L-histidine:D-mannose Npi-phosphotransferase | 39.8 | 122 |
| [4.2.1.30] glycerol hydro-lyase (3-hydroxypropanal-forming) | 39.8 | 64 |
| [3.4.17.19] Carboxypeptidase Taq | 39.9 | 122 |
| [2.7.1.189] ATP:(S)-4,5-dihydroxypentane-2,3-dione 5-phosphotransferase | 39.9 | 72 |
| [3.2.1.46] D-galactosyl-N-acylsphingosine galactohydrolase | 63.3 | 65 |
Compound Produced
This is reflected in compound produced. With D-Galactose being the only high – matching enzyme [3.2.1.46] D-galactosyl-N-acylsphingosine galactohydrolase above,
| CPID | Compound | percentile | obs |
| 924 | Ferrocytochrome | 36.9 | 63 |
| 22336 | Reduced hydrogenase | 38.3 | 121 |
| 250 | Pyridoxal | 38.6 | 98 |
| 15767 | 4-(L-gamma-Glutamylamino)butanoate | 38.7 | 63 |
| 16699 | 2-O-(6-Phospho-alpha-mannosyl)-D-glycerate | 38.7 | 94 |
| 1113 | D-Galactose 6-phosphate | 39.2 | 118 |
| 196 | 2,3-Dihydroxybenzoate | 39.3 | 73 |
| 4575 | (4R,5S)-4,5,6-Trihydroxy-2,3-dioxohexanoate | 39.5 | 72 |
| 615 | Protein histidine | 39.6 | 122 |
| 666 | LL-2,6-Diaminoheptanedioate | 39.7 | 122 |
| 1097 | D-Tagatose 6-phosphate | 39.8 | 115 |
| 20890 | D-Glucaro-1,5-lactone | 39.9 | 61 |
| 20889 | D-Galactaro-1,5-lactone | 39.9 | 61 |
| 124 | D-Galactose | 63.5 | 65 |
Compounds Consumed
As above for production and for bacteria, we have just one item with a high percentile and many with a low percentile.
| CompoundName | percentile | Obs |
| Galactosylceramide | 63.9 | 65 |
| 5-Oxoproline | 39.9 | 120 |
| Hydrogen selenide | 39.9 | 122 |
| 3-Dehydro-L-gulonate | 39.9 | 72 |
| Reduced riboflavin | 39.8 | 50 |
| Protein N(pi)-phospho-L-histidine | 39.6 | 122 |
| 5-Aminopentanoate | 39.6 | 106 |
| [SoxY protein]-S-disulfanyl-L-cysteine | 39.6 | 43 |
| (L-Seryl)adenylate | 39.6 | 60 |
| (2S,3S)-2,3-Dihydro-2,3-dihydroxybenzoate | 39.6 | 73 |
| 6-Phospho-beta-D-galactoside | 39.5 | 117 |
| Sucrose | 39.4 | 122 |
| Hippurate | 39.4 | 63 |
| alpha-D-Galactose 1-phosphate | 39.4 | 122 |
| 2-Hydroxy-3-keto-5-methylthiopentenyl-1-phosphate | 39.4 | 52 |
| D-Galactose 6-phosphate | 39.3 | 115 |
| 4-Aminobutyraldehyde | 39.3 | 47 |
| D-Xylulose 5-phosphate | 39.1 | 122 |
| Benzene-1,2,4-triol | 39 | 43 |
| L-Arabinonate | 38.6 | 52 |
| Pyridoxine | 38.6 | 98 |
| Salicylate | 38.3 | 44 |
| dTDP-4-acetamido-4,6-dideoxy-alpha-D-galactose | 38.3 | 60 |
| Oxidized hydrogenase | 38.3 | 121 |
| 3,4-Dihydroxyphenylacetate | 38.1 | 47 |
| Thymine | 37.8 | 45 |
| FMN-N5-peroxide | 37.8 | 45 |
| FMN-N5-oxide | 37.8 | 45 |
| Ferricytochrome | 37.4 | 63 |
| Hexadecenoyl-[acyl-carrier protein] | 37.2 | 47 |
| Protein lysine | 37.1 | 120 |
| 2-O-(alpha-D-Glucopyranosyl)-D-glycerate | 37.1 | 54 |
| L-Rhamnonate | 36.7 | 46 |
| Geraniol | 36.4 | 49 |
| 4-(L-gamma-Glutamylamino)butanoate | 36.3 | 44 |
| D-Glyceraldehyde | 35.6 | 60 |
| 3-Hydroxybenzoate | 35.3 | 41 |
| (2R)-3-Sulfolactate | 34.1 | 43 |
| D-Glucaro-1,4-lactone | 33.3 | 43 |
| D-Galactaro-1,4-lactone | 33.3 | 43 |
Bottom Line
The is no obvious imbalance between compounds produced and consumed. Thus the best path is to use the bacteria identified at the start. A quick select has been added to the menus.
- There are a few things that could be factors, for example D-Galactaro-1,4-lactone is 33.3%ile for consumption and 39.9%ile for production. Unfortunately, the microbiome is not a closed system and whether any surplus accumulates would be too speculative.
3 thoughts on “Multiple Chemical Sensitivity (MCS) – A Cause Found?”
Comments are closed.