This is a note for software engineers out there.
I have moved on to rework the existing code. The processing is done on a dedicated server (32GB of memory, SSD drives for SQL Server. A little over 15,700,000 combinations of taxon needs to be made. Each combination needs to get all data on these taxons that are concurrent in any samples.
- I.e. All samples that have Taxon 123 and Taxon 432 being reported.
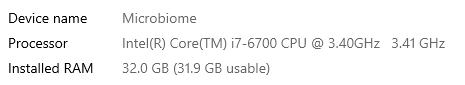
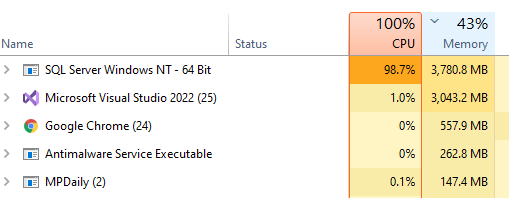
The computations are done using Parallel and Concurrent libraries. All indices have been tuned for this analysis. The SQL Server and the utility to calculate the associations are on the same server – so no latency or network impact.
I found that the CPU temperatures exceed the maximum recommended for the CPU chip.
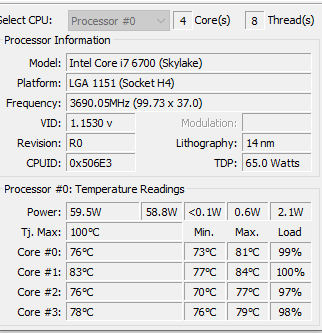
2022-03-27 20:12:08 Bacteria2Bacteria - 15,717,260 Items to Inspect 2022-03-29 22:48:34 Bacteria2Bacteria - 15,717,260 Items Inspected 2022-03-29 22:48:47 805,770 Items found
Bottom Line
This analysis likely qualifies for the “big data” label. So why is it worth it? The answer is simple, for some bacteria we have no information on what increases or decreases it. If we know which bacteria is associated with the bacteria growth or reduction, then we can synthesize modifiers of these bacteria that we lack information on.
This has not been implemented in suggestions (and will likely be available at the nerd level when it is).
This is very much interesting. Can I contribute in any way please.
Thanks
Sudhakar
Yes, you can look for alternative methodologies of computing associations.
The data is available at:http://citizenscience.microbiomeprescription.com/
At present we have 417,075 associations
Here’s the breakdown by R2
Assoc R2
11991 0.15
86991 0.2
92967 0.25
66248 0.3
47790 0.35
33855 0.4
23654 0.45
16960 0.5
11286 0.55
7734 0.6
5498 0.65
3363 0.7
2386 0.75
1576 0.8
1090 0.85
722 0.9
2464 0.95
500 1