A reader contacted me about a disagreement and the cause was a bug in the code for Kaltoft-Moltrup — subsequently fixed. This post looks at the bacteria selected by each for similarities and differences — so people can better understand the difference (which is a little abstract).
I am going to use one the demo samples from BiomeSight (BiomeSight:2019-06-10 Self).
- Extreme 3% picked 29 bacteria
- KM picked 24 bacteria
I sorted their selections below in alphabetical order, 13 are in common (just over 50% of the KM choices).
| Kaltoft-Moltrup | Extreme 3% |
| Actinomyces : Too High | Actinomyces : Too High |
| Actinomyces naturae : Too High | |
| Anaerofilum : Too High | |
| Actinomycetaceae : Too High | |
| Bacillales Family X. Incertae Sedis : Too High | Bacillales Family X. Incertae Sedis : Too High |
| Bacteroides cellulosilyticus : Too High | |
| Bacteroides denticanum : Too High | |
| Bacteroides dorei : Too Low | |
| Bacteroides intestinalis : Too Low | Bacteroides intestinalis : Too Low |
| Bacteroides rodentium : Too High | Bacteroides rodentium : Too High |
| Bacteroides sartorii : Too High | Bacteroides sartorii : Too High |
| Bacteroides thetaiotaomicron : Too High | |
| Bacteroides vulgatus : Too Low | Bacteroides vulgatus : Too Low |
| Blautia : Too High | |
| Blautia obeum : Too Low | Blautia obeum : Too Low |
| Brochothrix : Too High | |
| Brochothrix thermosphacta : Too High | |
| Chitinophagaceae : Too High | |
| Clostridium paradoxum : Too High | Clostridium paradoxum : Too High |
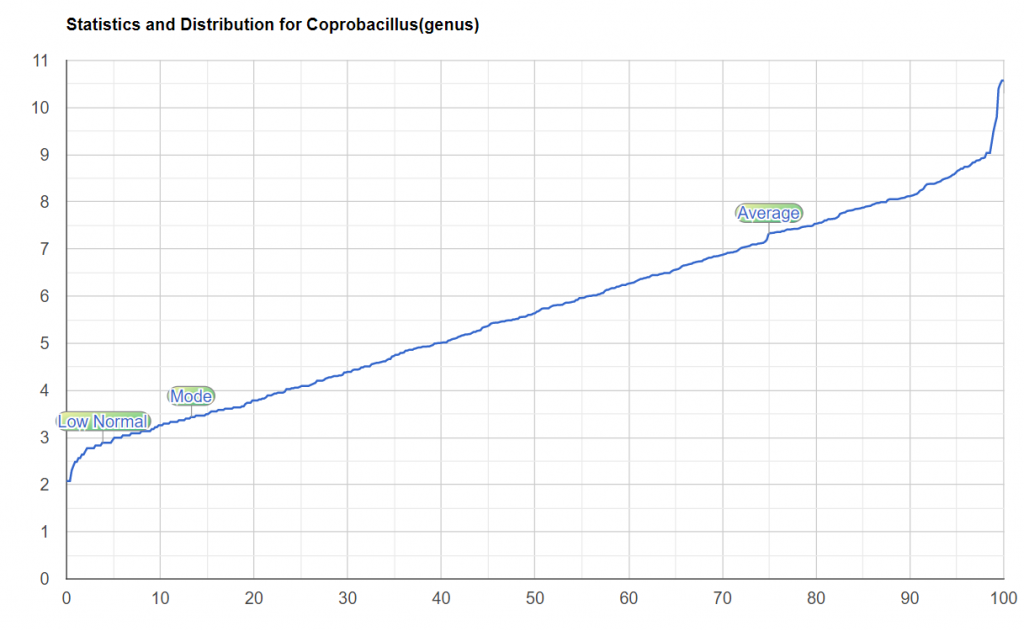
| Coprobacillus : Too High | |
| Coprococcus : Too High | Coprococcus : Too High |
| cunicula : Too Low | |
| Dehalogenimonas : Too High | |
| Desulfovibrio vietnamensis : Too Low | |
| Johnsonella : Too High | Johnsonella : Too High |
| Johnsonella ignava : Too High | Johnsonella ignava : Too High |
| Lachnospira : Too High | |
| Lactococcus : Too High | Lactococcus : Too High |
| Leuconostoc : Too High | |
| Listeriaceae : Too High | |
| Micrococcaceae : Too High | |
| Oscillospira : Too High | |
| Prevotellaceae : Too Low | |
| Streptococcaceae : Too High | |
| Streptococcus vestibularis : Too High | Streptococcus vestibularis : Too High |
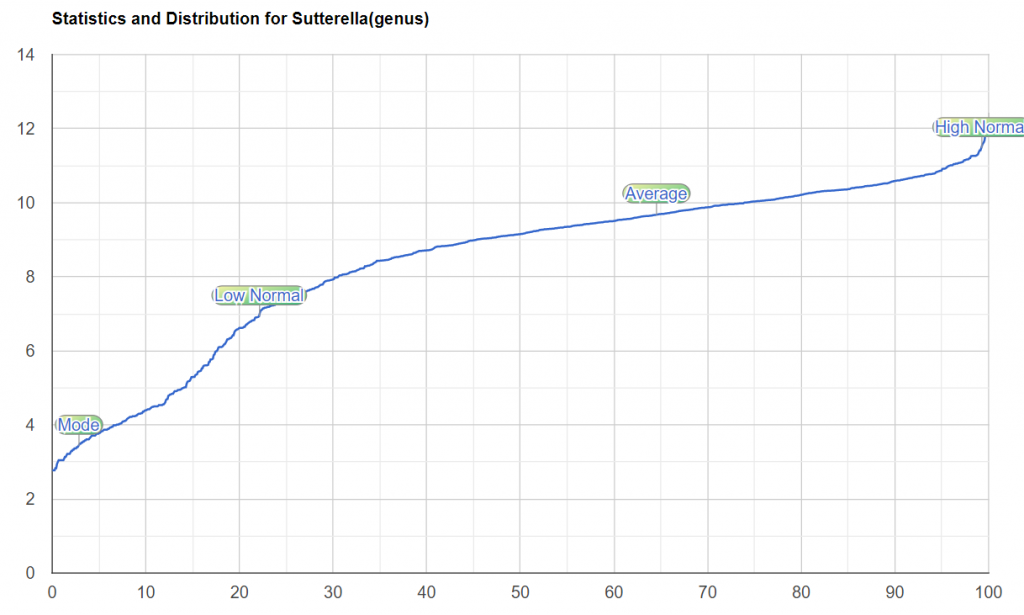
| Sutterella : Too Low | |
| Syntrophobacteraceae : Too High | |
| Tetragenococcus : Too High | |
| Thiothrix : Too High | |
| Turicibacter sanguinis : Too Low |
Looking at a chart of Prevotellaceae, we see that KM low is 2.25%, thus be this sample being between 2.25 and 3 resulted it being excluded on one and included on another. For Listeriaceae : Too High, KM used 95.6% instead of 97%.
For Sutterella, KM uses 22% for low, hence it included. This is reasonable because there is a distinctive drop off around that!
For Coprobacillus, it looks like I need to do some adjustments of the KM, a chunk of unusual data caused a “step” that incorrectly triggered the high computation.
Bottom Line
We have good overlap with the differences being due to the curves being different. With the extreme 3% approach, we are insensitive to the difference of shapes. With KM we are sensitive (and some parameters to the algorithm needs a little adjustment).
2 thoughts on “Comparing Extreme 3% to Kaltoft-Moltrup Selection”
Comments are closed.