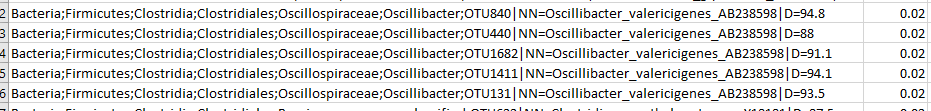
The report file reports only at the strain level, no genus or family levels are given. These total sums up to 100%. The smallest resolution appear to be 0.02% That is 1 in 5,000 bacteria. This is a lot lower resolution than other providers ( 1 in 160,000 is seem in some other reports with a good sample). There is something odd about a large number of bacteria being at 0.02 or 0.04 percent.
It appears that FASTQ downloads from them (alleged to be available if requested) is the prefered way to get better data.
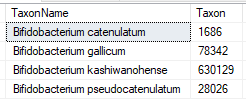
One bacteria was listed as:”(Bifidobacterium catenulatum/Bifidobacterium gallicum/Bifidobacterium kashiwanohense/Bifidobacterium pseudocatenulatum)”
which is with more current tests are 4 different strains.
Bottom Line: Won’t Do
There are too many problems with the data. I have spent almost an entire day fighting it. If they provide a FASTQ file, I have unload for those processed through SequentiaBiotech web site.
To use their CSVs:
- They must provide the official Taxon Numbers in the Excel File
- They must provide the full hierarchy with numbers at each level
Without those, their data will pollute the existing contributed base too much. There are no acceptable kludge arounds for these defects.
I would guess that 0.02% is one organism found, and 0.04% is two organisms found.