I started Microbiome Prescription site using data uploads from ubiome, a firm that was founded by a crowdfunding campaign, went to venture capitalists, and went unethical due to pressure from venture capitalists and died. I received over 800 samples processed by ubiome.
Readers started to request the microbiome reports to be processed on the Microbiome Prescription site and I started adding them according to constraints of the reports available. BiomeSight.com, a UK firm, has been the most cooperative. We worked together to allow automatic transfers directly from their web site to the Microbiome Prescription site by using a API.
Xenogene | Metagenómica y Biología Molecular Reports were shared to me. I found that I could do an accurate extract from one of the reports they made available to users. The result was that they became the most comprehensive report as see by the statistics below

There were few uploads because of their higher costs. The report that I used is shown below.

They Changed Their Report
Recently I have had two people trying to upload their reports. The report was different than the above. I asked them to contact Xenogene to get the above; they were not successful. I examined their reports (they were several years apart), and found two different formats, as shown below:
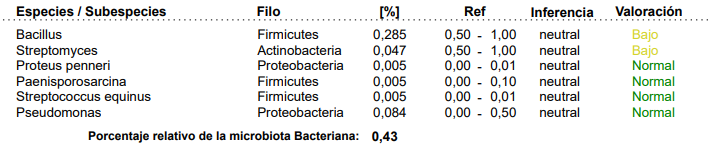
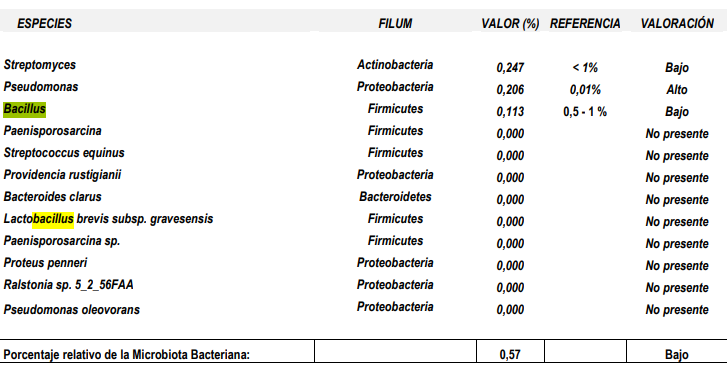
While both give the same information, the structure of the page was different. The report do not give the hierarchy, for example, Eubacteriaceae was found in neither report. I looked for Blautia and could find species and strains — but no total, so you cannot apply Dr Jason Hawrelak criteria.
Summing up all of the species and strains under Blautia does not give a correct total, in some cases the Blautia total will be 2x higher. Why, because many strains and species has not received names and / or “fingerprints”.
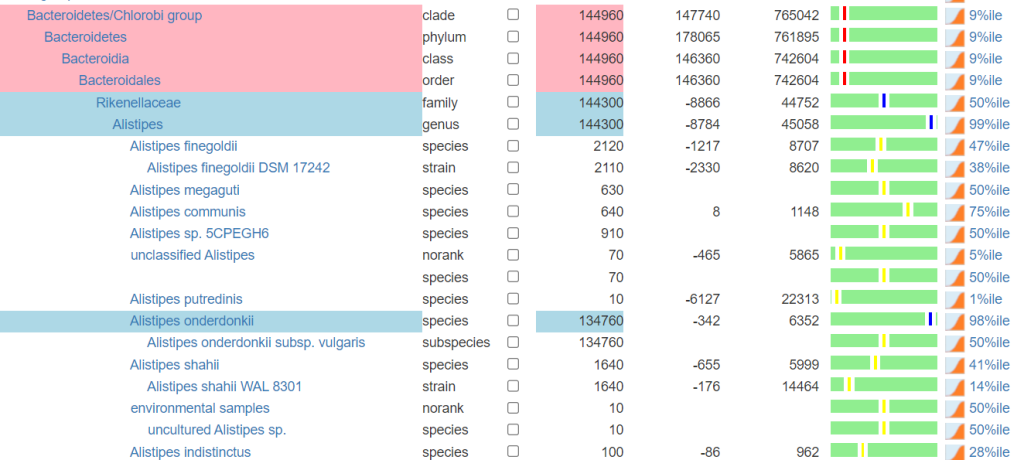
Bottom line: I no longer recommend this lab
Despite these issues, I have updated the import to support both of the above PDF formats and synthesize all of the missing layers of the bacteria hierarchy. The new import should be on line by November 13th.
WARNING: the genus level and above may often be low because of the total synthesis.
I did extract their recommended ranges and added it as an option:
Citizen Science Action
I would suggest emailing them and asking them if they make a CSV file available of the results (including the bacteria NCBI taxon number), if so, can you get a sample. If you do not get a positive results, do a return email asking you to be informed if they change and indicating that you are going to use Ombre or Biomesight instead…… The risk of loosing customers can often change business practice.
Microbiome Activism Please!!
This applies not only to Xenogene but
- Bioscreen (cfu/gm)
- Biovis Microbiome Plus (cfu/g)
- DayTwo
- Diagnostic Solution GI-Map (cfu/gm)
- GanzImmun Diagnostic A6 (cfu/gm)
- GanzImmun Diagnostics AG Befundbericht
- Genova Gi Effects (cfu/g)
- Genova Parasitology (cfu/g)
- GI EcologiX (Invivo)
- GI360 Stool (UK)
- Gut Zoomer (vibrant-wellness)
- InVitaLab (cfu/gm)
- Kyber Kompakt (cfu/g)
- Medivere: Darm Mikrobiom Stuhltest (16s limited)
- Medivere: Darn Magen Diagnostik (16s Limited)
- Medivere: Gesundsheitscheck Darm (16s Limited)
- Metagenomics Stool (De Meirleir) (16s Limited)
- Smart Gut (ubiome 16s – Limited Taxonomy)
- Verisana (cfu/ml) aka (kbe/ml)
- Viome (No objective measures)
and also these lacking NCBI taxon numbers
In Australia, after being a loyal Ubiome customer since you began the marvellous Microbiome Prescription, Ubiome puzzlingly wrote to me to say that Australia no longer permitted human specimens to be exported, (untrue) not saying that UBiome itself had collapsed. so it good to get your above explanation. We sent a sample off to the UK Biomesight for the first time in October. But after a month they’ve written to me saying that our sample had not arrived! I had no tracking records. The great news is that in the same email, they offered to send a replacement free of charge. I said I’d report it to you- and will report if it does indeed turn up.