David Morrison requested this feature. The FastQ file is produced by the physical lab machine. This file is then pushed thru software to produce a list of taxonomies. Different 16s retail providers use different software and as a result – different reports. For back ground see this “Taxonomy Nightmare before Christmas” post.
Walkthru
This feature is available only for those that sign on securely (i.e. request a login link to be sent by email).
On the samples page you will see some new buttons
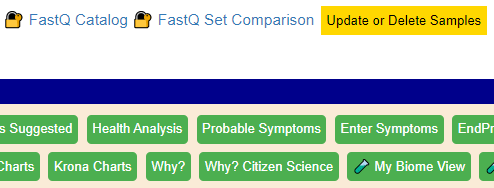
Define FastQ files
Simply Click [FastQ Catalog] and create records for each FastQ file you are going to work with.
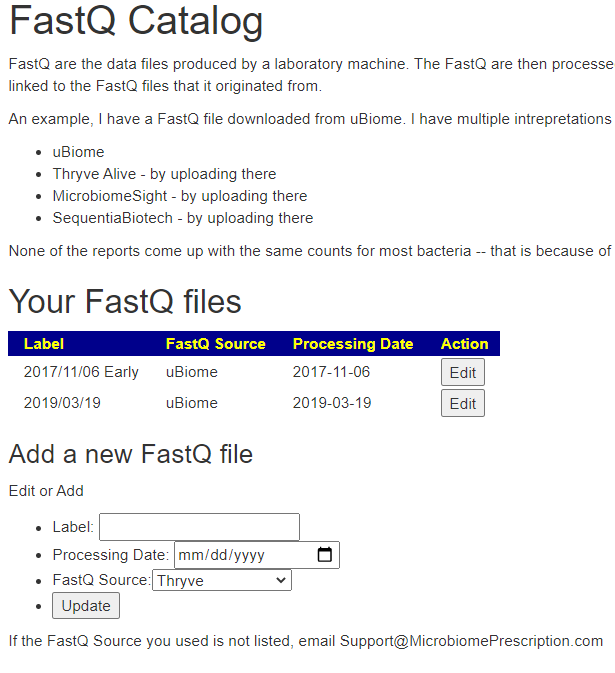
Identifying sample with FastQ file
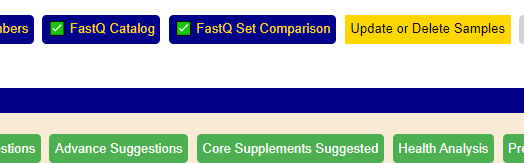
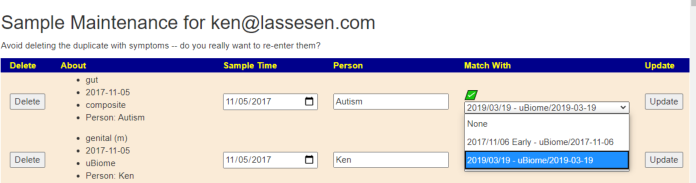
Return to the Samples page, and click [Update of Delete Samples]
Comparison
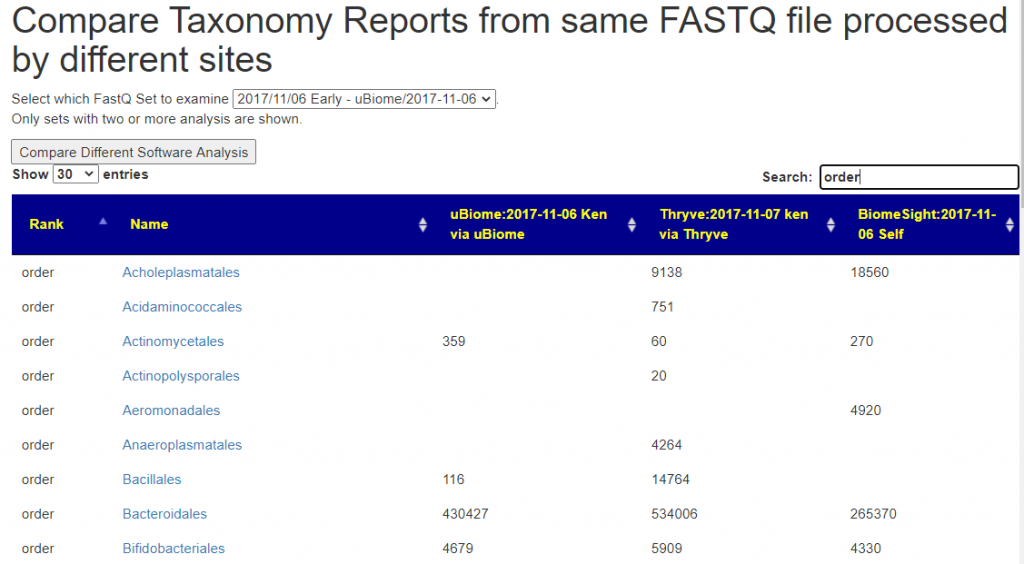
You need at least 2 sample to be linked to a FastQ file. Returning to the Samples page, click the [FastQ Set Comparison] button


You will now see the results reported. Some taxonomy will be very similar and others very different.
What’s next?
Once sufficient users link their files, we can start doing the various analysis that David was asking for.
Recent Comments