I just pushed out an update on http://microbiomeprescription.azurewebsites.net/ that may help you understand what various prescription, over the counter and some supplements may be doing to your microbiome.
Select any of the links highlighted below
The next page will show some choices at the top:
Compare Impact
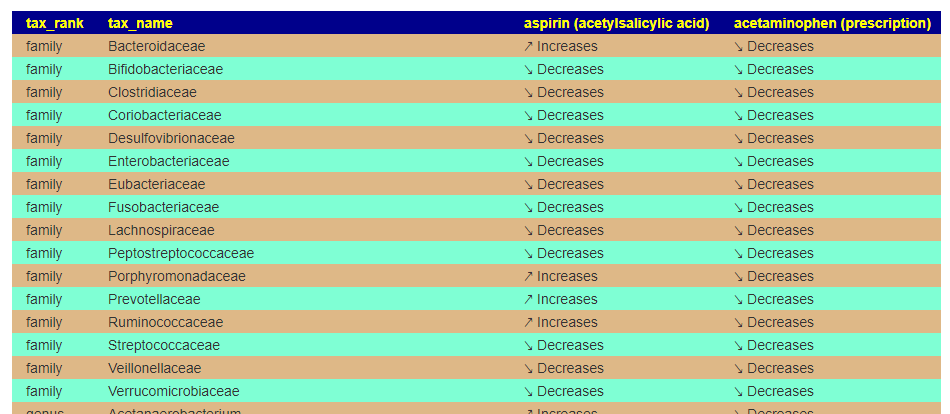
This is intended to allow you to better choice between alternatives – for example Aspirin versus Paracetamol (acetaminophen). I am sure people will find more uses for it.
The process is simple, search for each item, and put a check beside it. Select the Compare Impact radio button and then click the submit button below it.
This will take you to a page listing the impact side by side. In this case we seel that their impacts are similar, but different on a few items. At the family level there are a few differences
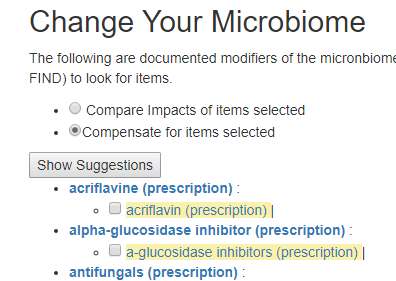
Compensate
This is intended when you are prescribed drugs to treat some conditions and wish to reduce the impact on the microbiome by counteracting the drug or drugs impact on the microbiome.
For this example, we pick lovastatin (a statin), Famotidine (Pepcid AC).
We may wish to first see how much impact they have together (do they reinforce or counteract each other)
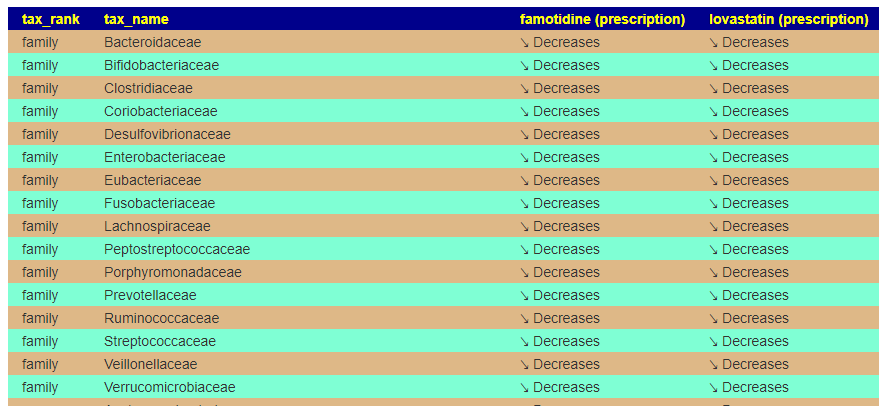
Just pressing back, and changing radio buttons, and submit produces suggestions.

The suggestions are done by creating a virtual microbiome report based on the above shifts and running that through our AI engine.
The suggestion page is the new format with the long lists hidden until you ask to see them.
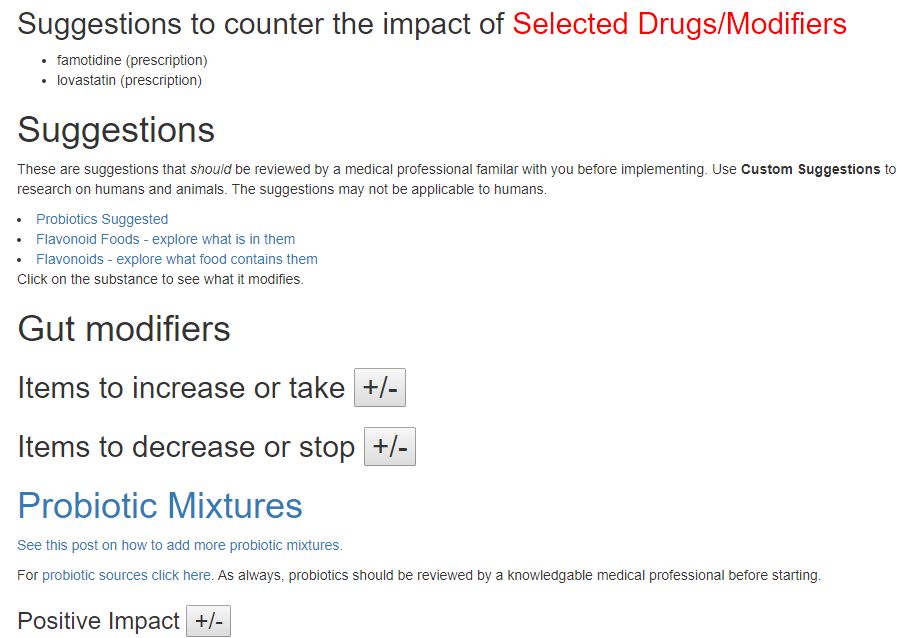

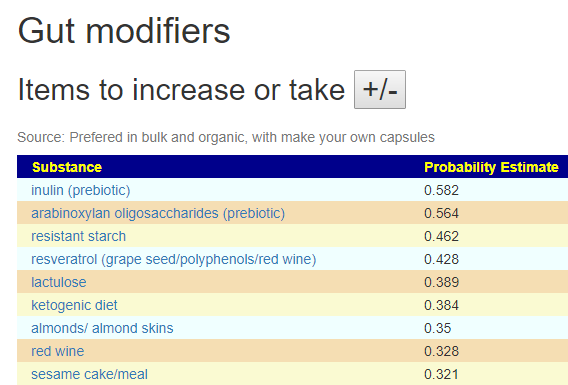
The Take or Avoid list is defaulted to 100 items (which is one reason that I toggle visibility). Remember – none of these items are guaranteed to work, nor do you need to take all of them. Each item increases your odds…
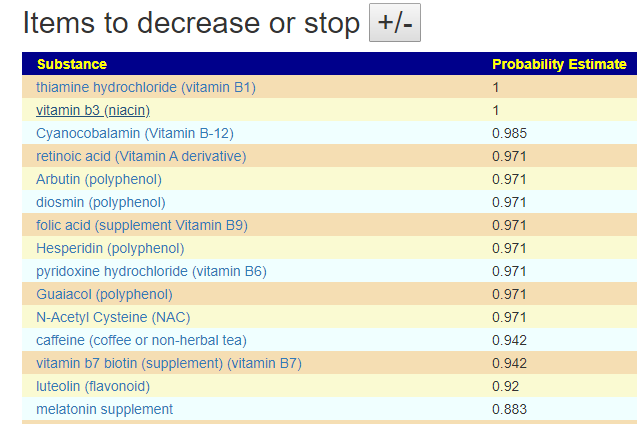
The avoid list values are a lot higher, and thus you may wish by reducing any of these items that you are taking.
Recent Comments