A colleague, DM, in Australia forwarded A core microbiome signature as an indicator of health [2024] to me this morning. It uses the concept of guilds. I responded with “that is very close to my underlying model. I.e. the net metabolites produced from these ‘guilds'”. An image from this paper is below.
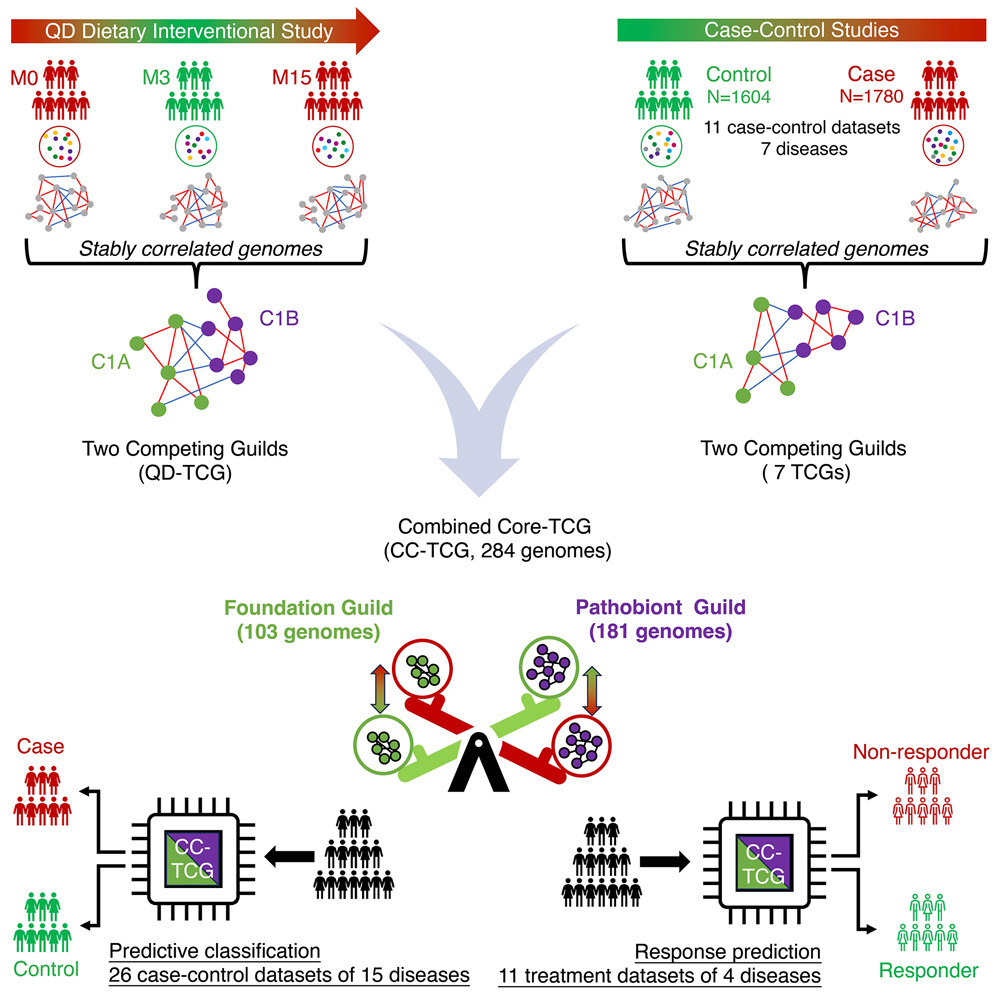
The concept of guilds was advocated by Guild-based analysis for understanding gut microbiome in human health and diseases [2021] and described as:
Translating the concept of “guild” to the study of gut microbiota, we redefine guild as a group of bacteria that show consistent co-abundant behavior and likely to work together to contribute to the same ecological function.
I am not a life scientist, I am a Operation Research/Statistician/Mathematician person. I have taught different life sciences at high school and college levels — so I am able to read life science papers well. Professionally, I have done a wide variety of Artificial Intelligence for decades as well as world class software development with many papers published by Microsoft (and a few patents). Life sciences tend to be slow (or glacial) in adapting methodologies well practiced in other disciplines.
My Core Insight
At the start of my journey into the microbiome, I saw inconsistent results in papers and quickly concluded that the diagnosis and symptoms were not the direct result of bacteria but likely the cross of DNA with the basket (to borrow a term from economics) of chemicals being produced by the bacteria. The inconsistency of taxonomical identification (see The taxonomy nightmare before Christmas…) added another level of fuzziness to it. Fuzziness is very familiar to old time AI and statistics folks, with skill sets to address it.
What I started referring to as chemicals, is more correctly named metabolites and enzymes. I developed a methodology using data from KEGG: Kyoto Encyclopedia of Genes and Genomes, to estimate relative levels from microbiome samples and then did the logical step of identifying the bacteria that would normalize them. The bacteria was limited to probiotics available retail somewhere in the world.
The results were the specific bacteria followed by the retail brands.

At this point, I need to emphasis that my focus is on the common person suffering from symptoms and conditions and not the academic world.
A Better Solution is Available
People familiar with my site knows that I use the Monte Carlo model to generate suggestions. Using multiple criteria to filter the bacteria of interest, apply slightly different algorithms, and produce suggestions from the results to maximize the likelihood of them helping. The core algorithm cross validates with 86% correctness ( see Suggestions Cross Validation using PubMed), so this approach has been very successful for individuals reducing symptoms (or having them disappear).
Above, I cited “estimate relative levels”, with current technologies and the appropriate lab (for example Clinical Microbiomics ) we can eliminate or reduce the “estimate” aspect. The next aspect is to extend the microbiome adjustment beyond probiotics. This is very do-able with a background in Mixed-Integer Programming (i.e. Operations Research skill set).
The Mixed-Integer Linear Programming problem is selecting modifiers that will cause the existing bacteria to be adjusted to normalize the metabolites and enzymes. If some metabolites or enzymes are known to be particularly significant for a symptom or condition, then you assign a higher weight to those.
With modern computers. this is a manageable problem. You end up with a three dimensional spread sheets:
- Likely 5-8000 bacteria from a microbiome test
- Likely 8-12000 metabolites and compounds
- Likely 2000 modifiers (we are limited to compounds that we know influence bacteria from studies)
The result is 8000 x 12000 x 2000 = 192 billion cells in our spread sheet. A well provisioned computer can easily handle this in memory. For example: Dell: Precision T7500 workstations or BOXX Technologies: APEXX S-Class workstations. Windows 10 Enterprise supports up to 2000 GB of memory.
It should be noted that each item likely have acceptable ranges. For example, excessive methane production could result in a SIBO diagnosis. The old adage, Do no harm, applies.
There are more dimensions that could be added, for example “cross reactivity between bacteria strains”, but the available knowledge there is quite limited. Similarly, we could include functions instead of assuming linearity — again, the available knowledge there is quite limited.
My Stumbling Blocks
Why have I not already done this? The answer is that I have done a reduced set of the approach described above. The optimization is time consuming and not suitable for immediate suggestions on a web site. Doing it on “the cloud” will result in some very nasty monthly bills. As stated above, my focus is on the common person with challenges. Part of this focus is making the solution explainable to the common person. “Taking probiotics to better balance the enzymes and metabolites” is understandable. I can walk a person through the mathematics of each suggestion and thus get “buy-in”. Explaining the Mixed-Integer-Programming solution above is very similar to try explaining what the random forest algorithm is.
Bottom Line: It is possible to significantly improve suggestions to modify the microbiome, there are a few challenges.
Recent Comments