A reader sent an email with some good questions. He is pleased with the results. Looking at repeat users of Microbiome Prescription (tried suggestions and came back within a year for more suggestion) is almost 69%. This suggests happy users.

That system works great!
- Could you explain the main differences between the OLD UI and NEW UI? Sometimes the data doesn’t seem to match up well, and I’m unsure which one I should use.
- I’d like to understand the symptoms sections better, as they look very different in both UIs. The old UI symptoms make much more sense for me.
- I have a Biomesight test for a friend with many gut symptoms, but when I analyze the data, I’m not seeing much in terms of actionable recommendations for things to add or remove. I do see a little more in terms of statistical significance in the OLD UI. What would be the most accurate way to read the data in a case like this?
- I primarily use the full consensus reporting feature in the database. Are there any advanced features or sections you think I should familiarize myself with?
- What is the main difference between symptoms and medical conditions in the database? My understanding is that medical conditions are more supported by proven data, while symptoms are based more on self-reporting. Is that correct?
- If possible, I’d love to see your workflow for analyzing a test.
Could you explain the main differences between the OLD UI and NEW UI? Sometimes the data doesn’t seem to match up well, and I’m unsure which one I should use.
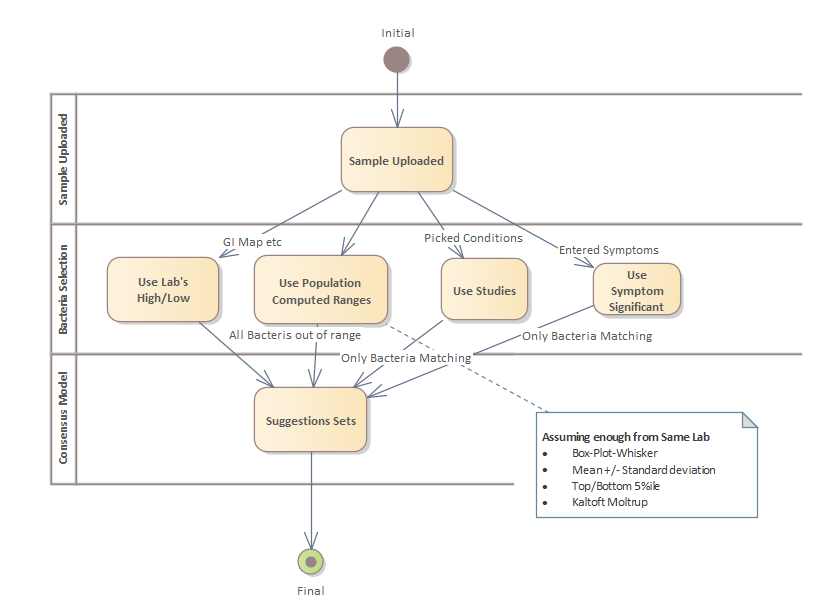
The site evolves as I keep getting better insight on data and different ways of getting statistically significant data. In general, when I get a new insight it is added as a new feature while keeping the older approaches. The older approaches appear to work well, but I want to keep pressing forward finding “better ways”. Actually, the way may not be better for everyone — rather better for some cases. It is the classic “no algorithm works for every one”.
Using Monte Carlo Model that builds consensus suggestions, my hope is that these various approaches will net better suggestions.
- I avoid dropping methods. It upsets some people. Also these older methods work well for some.
Suggestion: Use both and work with the Consensus
I’d like to understand the symptoms sections better, as they look very different in both UIs. The old UI symptoms make much more sense for me.
At present there are at least three different ways of forecasting symptoms. Most of the methods pick slightly different sets of bacteria with different weights. Forecasting symptoms depends on which regression / modelling is used. Some examples:
- Over and Under Represented for Symptoms by Lab
- Symptom Association Studies which can be based on many approaches
- Difference of Average or Difference of Median
- Chi-square values between top and bottom X%iles
- etc
I have in my backlog to test each of these methods to evaluate their ability forecast symptoms. This also require tuning each of these to try to get the best accuracy in forecasting. That is likely at least a month of work (once I get the cycles).
In short, different methods were tried to detect statistical significance using both parametric and non-parametric methods. When there were a sufficient number of bacteria found significant, then a forecaster is built.
When I get time to do comparison before forecasting accuracy, the number of choices will likely be reduced.
I have a Biomesight test for a friend with many gut symptoms, but when I analyze the data, I’m not seeing much in terms of actionable recommendations for things to add or remove. I do see a little more in terms of statistical significance in the OLD UI. What would be the most accurate way to read the data in a case like this?
IMHO, the most accurate is checking the symptoms they have and use that for suggestions. It is the most likely way to pick the significant bacteria to focus on.
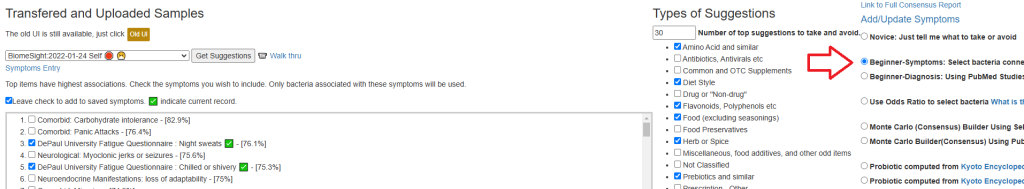
I usually use the metabolites and enzymes approach to select probiotics. Typically this will be the same probiotics in suggestions but in a different order. I give the probiotics suggested based on KEGG data a higher value because the suggestions above are based on what has been studied (which tends to be erratic). The KEGG data is based on the DNA/RNA of the microbiome and far less sensitive to what has been studied in clinical studies.
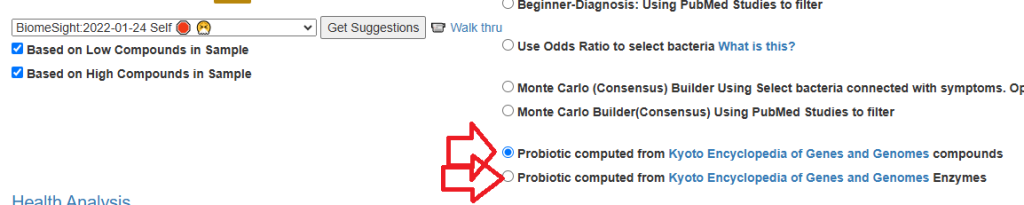
I primarily use the full consensus reporting feature in the database. Are there any advanced features or sections you think I should familiarize myself with?
Nothing at the moment, On the [Changing Your Microbiome] under “Suggestions for building general consensus”. These are the four most promising methods. New methods will likely be added at the bottom of this list as they are added.

What is the main difference between symptoms and medical conditions in the database? My understanding is that medical conditions are more supported by proven data, while symptoms are based more on self-reporting. Is that correct?
Medical conditions are those reported in the literature — unfortunately every study uses different processing. If the same study samples processed through a different process, different bacteria will be found significant (See The taxonomy nightmare before Christmas…). These are “best efforts” selection when we do not have sufficient data for a condition or symptoms for the specific processing lab that you are using.
The “inhouse” associations are always done using data from the same processing lab, so the identification of the “lab named bacteria” are consistent. This is the most likely to pick the right bacteria (according to the lab). One major difference is that Medical conditions are often based on 30-60 samples alone. For Biomesight data, we often have 600 samples and thus better ability to identify.
If possible, I’d love to see your workflow for analyzing a test.
Recent Comments