As a reminder, Microbiome Prescription is a “best efforts” site. We do the best that we can with the data that is available.
Request from Reader
Hello Ken,
I would kindly like to ask your opinion on this. I did three BiomeSight tests in three years, then one Xenogene. The Xenogene and last BiomeSight were 10 days apart.
BiomeSight was telling me for three years that my butyrate producers were awesome and my F.Prau was great. Then Xenogene told me the exact opposite. BiomeSight results being so good for so long, I always assumed I have no problem in the microbiome so I kept my diet (15g of fiber at max, lots of meat, veggiest mainly potatoes – no 30-40 different veg/fruit per week). Butyrate producers and F.Prau this high on this diet is a little hard to believe, so I came to the conclusion that BiomeSight is completely off, at least for me.
I remember the Taxonomic nightmare article you wrote and I understand that I cant directly compare two test providers. But when one test tells you that your microbiome is a rockstar and the other tells you its a zombie, its hard to see the usefulness of biomeSight testing. I kindof hope there is some magic that Im forgetting and the biomeSight tests will not prove to be a waste of time and money.
The Xenogene values are percentages of bacteria only (i.e. # F. Prau / # total bacteria), so its “the same thing” as in biomeSight results. Just to be clear that its not # F.Prau / (# bacteria + # protozoa + # archaea + # fungi).

Explanation
Numbers always need to be interpreted against reference ranges.
Standard Lab Reference Ranges
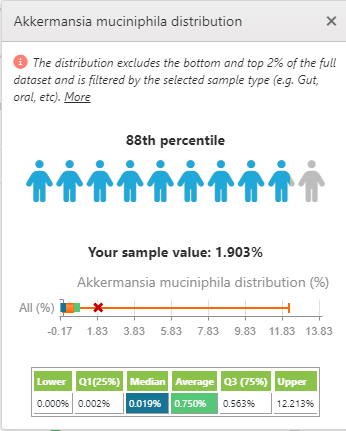
If you go to the Akkermansia reference page, we see the following:
| Lab | Mean | Standard Deviation | Shift in S.D. |
| Xenogene | 0.693 % | 1.074 % | – 0.9 |
| Biomesight | 1.393 % | 4.111 % | +.83 |
In other words, both test results were within 1 standard deviation of the mean – that will usually be interpreted as in the normal/reference range. The formula is easy:
(Your Value - Mean) / Standard Deviation
You can do this for each of the bacteria in your report. If the resulting value is between -1.6 and +1.6, you are clearly in the reference ranges.
Lab Provided Ranges
Xenogene provides ranges — this means that over 70% of xenogene files uploaded to MP are below the reference range.
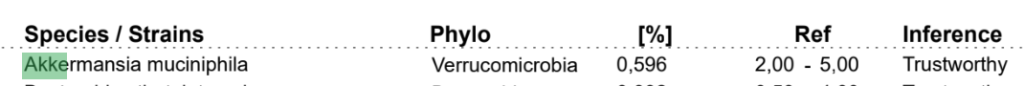
Biomesight reference ranges are below
Every value is within the reference ranges. We have a disagreement.
Dr. Jason Hawrelak Recommendations
His ranges are 1-5% for Akkermansia. These are much less than Biomesight and bigger than xenogene.
User Feedback: “Dr. Hawrelak’s ranges – how can Dr. Hawrelak have a general range on some bacterium w/o stating which lab he uses for this? Considering all labs report different numbers due to the taxonomy nightmare, I don’t understand how there can be “one range to rule them all”. “
I agree totally, some labs cites him as an authority because they lack the skilled resources to determine their lab specific ranges, Often I have seen ranges from a published studied applied to numbers from a totally different processing pipeline – when challenged they cite “it’s an authority“. Some more readings:
As a FYI: I include his ranges because people have requested it. I provide choices and not judgements.
Microbiome Prescription Ranges
We do not have enough data to independently compute xenogene. Xenogene samples are part of “Other Labs”. The Kaltoft-Møldrup ranges are:
- Other Labs: 0 – 7%
- Biomesight: 0 -9.6%
Similarly using BoxPlot methodology,
- Other Labs: 0 – 3.8%
- Biomesight: 0 -1.7%
Note that Zero (0) is in range for many of these.
The Symptoms Factor
Identifying bacteria associated with symptoms depends on the number of samples uploaded with annotated symptoms. We do not have sufficient results with Xenogene, we do have sufficient for Biomesight with some 289 associations at present. Note this is pattern matching. “It has the ears of a German Shepard, it has a double coat of a German Shepard, it eyes color matches a German Shepard…etc. ” It may be German Shepard or it may be a Wolf or a Welsh Pembroke Corgi
For the latest Biomesight, we see a lot of matches to existing patterns

For Xenogene (which uses “Other Labs”) we have less and weaker matched

If you want to include your symptom in the suggestions report, biomesight is a better choice.
Eubiosis
Eubiosis is not very comparable, because Xenogene is mixed with all of the other odd labs. We have just 19 xenogene, not sufficient to do that much data, We have 61 samples from Thorne – same issue.
Follow up Questions
So my question was how can BiomeSight tell me my butyrate producers are 60% when its highly unlikely, just as with F.Prau being 19%. And the answers that I could imagine getting would be
- you having 60% butyrate producers is the result of taxonomic hell, in some cases the positive measurement errors can add up resulting in a hugely overestimated sum – judging the SCFA producers in the biomeSight report is unreliable and should not be taken into consideration at all
- OR the only relevant marker for abundance of SCFA producers is stool pH or SCFA measured in stool – use that instead of the SCFA producers % reported by ANY test provider
- OR something else if the former two are nonsense
Butyrate evaluation is a good illustration minefield. Some observations:
- Labs will usually qualify with “It is important to note that this is not a measure of these metabolites found in the stool sample.“
- One lab totals 9 Genus and 1 species.
- Biomesight appears to total 21 genus
- Another totals: 43 species
- Microbiome Prescription is based on KEGG: Kyoto Encyclopedia of Genes and Genomes :
- 402 species that could produce butyrate
- 515 species that could consume butyrate
MP is the only one that appears to include consumers (thus getting a net amount). The choice of genus and species is often based on the depth of research that each lab does. MP is based on the genetics of the bacteria as sequenced and aggregated by KEGG. We do not know if the genetics are activated or not (epigenetics).
What is missing are studies comparing various estimates from bacteria against actual directly determined levels of butyrate and other metabolites…
Second Issue: Percentage of WHAT?
Many labs pull a magical number out of the air, typically if you are over this number you are Satisfactory, below Not Satisfactory. MP gives a percentile ranking against others samples using the same lab. If you are over 30%ile, I would deem it to not need work — but that is a personal judgement call, the numbers are there for you.
Bottom Line
Microbiome reports are full of uncertainty aka fuzzy data. Fuzzy data is not a strong selling point for businesses. Claiming accuracy and correctness is a great marketing ploy. If there are no legal/financial consequences of these dubious claims then they will typically be made by some.
I have seen some labs that started by “just reporting the facts/numbers” and then drift into interpretations because marketing studies found that would increase business.
Xenogene (and Thorne) is good because of what else it reports that is not reported on Biomesight which may be part of your health issues. For items that seem very high, you should do some research on them and if any medical conditions are reported/associated with them.
On the other side, we have a lot of samples with annotated symptoms for Biomesight and Ombre. This means that suggestions to modify your microbiome (especially if symptoms are used) are likely better.
My usual advice is simple:
- do one of Xenogene or Thorne to check for non bacteria issues (fungi, phages, etx)
- use Biomesight or Ombre for regular testing and getting suggestions.
Recent Comments