This is a series of samples from a long time suffer from Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). In looking at the samples, we must keep in mind two factors:
- Lab Read Quality. A low read quality reports much fewer bacteria than a high read quality.
- 1.8 reports 300, a high read quality can triple the bacteria counts selected.in a category
- This usually does not impact the broad stroke criteria such as Dr. Jason Hawrelak (JasonH)
- A sickness (COVID, Flu) or even vaccination will alter the microbiome and create the appearance of lost ground. This was seen and covered in the post below
My usual practice is compare the latest with the prior only. Trying to weave a dialog covering all prior tests is contra productive with going forward. What is past, is water under the bridge.
This reader had one major question — the suggestions seem to have changed a lot. That is the focus of this post. Have there been a dramatic change, or are the changes likely connected to a few bacteria dropping off the list and a few new ones being added.
Earlier post for this person
Person’s Subjective Report: “Symptom-wise and treatment-wise, I don’t have much to report that is new or an outlier to previous years of samples and treatments.”
Review over Multiple Samples
Data from the same lab (using Ombre data, Biomesight version is also available).
Compared to the prior sample, we see:
- Less extreme percentiles than prior sample
- Less rarely seen bacteria
- Less pathogens
- Less outside of:
- Outside Lab Range (+/- 1.96SD)
- Outside Box-Plot-Whiskers
- Outside Kaltoft-Møldrup
- No change using a multitude of 3rd party criteria.
- Less compounds at high levels, more compounds at low levels
General impression, there has been objective improvement
| Criteria | 4/18/2023 | 2/2/2023 | 11/1/2022 | 4/11/2022 | 1/11/2022 | 03/09/2021 | 5/27/2020 |
| Lab Read Quality | 4.8 | 5.3 | 8.6 | 15.3 | 1.8 | 2.9 | 4.3 |
| Bacteria Reported By Lab | 638 | 734 | 800 | 750 | 312 | 394 | 544 |
| Bacteria Over 99%ile | 4 | 14 | 12 | 3 | 9 | 3 | 4 |
| Bacteria Over 95%ile | 36 | 42 | 24 | 21 | 30 | 10 | 12 |
| Bacteria Over 90%ile | 56 | 79 | 46 | 49 | 46 | 25 | 36 |
| Bacteria Under 10%ile | 25 | 46 | 154 | 329 | 16 | 26 | 32 |
| Bacteria Under 5%ile | 10 | 21 | 54 | 287 | 10 | 15 | 11 |
| Bacteria Under 1%ile | 2 | 1 | 7 | 84 | 1 | 5 | 0 |
| Lab: Thryve | |||||||
| Rarely Seen 1% | 10 | 15 | 7 | 8 | 0 | 2 | 9 |
| Rarely Seen 5% | 36 | 61 | 58 | 70 | 2 | 4 | 18 |
| Pathogens | 27 | 34 | 32 | 30 | 21 | 26 | 30 |
| Outside Range from JasonH | 4 | 4 | 4 | 4 | 7 | 7 | 7 |
| Outside Range from Medivere | 13 | 13 | 19 | 19 | 14 | 14 | 14 |
| Outside Range from Metagenomics | 10 | 10 | 7 | 7 | 9 | 9 | 9 |
| Outside Range from MyBioma | 13 | 13 | 10 | 10 | 8 | 8 | 8 |
| Outside Range from Nirvana/CosmosId | 22 | 22 | 22 | 22 | 17 | 17 | 17 |
| Outside Range from XenoGene | 51 | 51 | 48 | 48 | 42 | 42 | 42 |
| Outside Lab Range (+/- 1.96SD) | 18 | 24 | 14 | 17 | 8 | 5 | 5 |
| Outside Box-Plot-Whiskers | 67 | 97 | 63 | 63 | 58 | 28 | 45 |
| Outside Kaltoft-Møldrup | 143 | 218 | 290 | 400 | 89 | 102 | 141 |
| Condition Est. Over 99%ile | 0 | 0 | 0 | 5 | 2 | 1 | 0 |
| Condition Est. Over 95%ile | 0 | 0 | 2 | 24 | 6 | 5 | 1 |
| Condition Est. Over 90%ile | 0 | 7 | 3 | 38 | 11 | 9 | 2 |
| Enzymes Over 99%ile | 13 | 47 | 44 | 4 | 1 | 41 | 56 |
| Enzymes Over 95%ile | 114 | 179 | 99 | 72 | 7 | 311 | 465 |
| Enzymes Over 90%ile | 469 | 266 | 242 | 152 | 54 | 683 | 512 |
| Enzymes Under 10%ile | 133 | 123 | 219 | 430 | 70 | 141 | 106 |
| Enzymes Under 5%ile | 58 | 47 | 108 | 310 | 47 | 88 | 53 |
| Enzymes Under 1%ile | 7 | 1 | 1 | 64 | 6 | 14 | 5 |
| Compounds Over 99%ile | 11 | 31 | 22 | 3 | 1 | 27 | 53 |
| Compounds Over 95%ile | 37 | 122 | 67 | 46 | 9 | 305 | 276 |
| Compounds Over 90%ile | 146 | 190 | 231 | 93 | 38 | 488 | 365 |
| Compounds Under 10%ile | 954 | 809 | 823 | 998 | 530 | 617 | 800 |
| Compounds Under 5%ile | 906 | 789 | 788 | 961 | 521 | 599 | 787 |
| Compounds Under 1%ile | 872 | 783 | 757 | 892 | 515 | 582 | 776 |
Comparing Suggestions
The reader noticed a significant change of suggestions between the samples. So I am going to document out how to verify or see the results. Also, remember that you can merge consensus from two different samples. Minor items like taking samples at different time of days, unusual food, significant dosages of herbs, probiotics or supplements within 48 hours of taking a sample can cause temporal shifts.
Step by step:
- Do “Just Give Me Suggestions” for 1st sample
- Click “For more technical details”
- Click Download, save the file
- Repeat for 2nd sample
- Open Excel or equivalent on the .csv file downloads
- Each sample will be in a difference instance of Excel
- Add a new work sheet to one
- Copy one set of data to the other
- Rename worksheets as “Current Consensus” and “Prior Consensus”
- Copy the Mid2 column to the first column in the Prior Consensus”
- Insert a blank column after the Mid2 column,
- Insert into this new blank column: =VLOOKUP(I2,’Prior Consensus’!A:J,2,FALSE)
- This brings the net value over.
- You can brink other values over by changing “2”
I have done a video below of the process.
Wait! This is more for your mobile phone users
Go to Multiple samples, then pick your samples as shown below
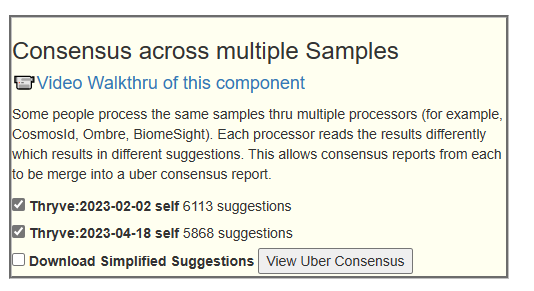
You will now see a summary at the top
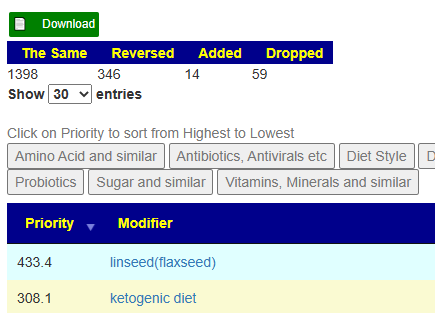
Bottom Line
I have not done further analysis etc. The reader has acquired those skills already. I have address only the issue of shifting suggestions. My best advice is simple: Do an uber consensus between the present and the last sample. The logic is simple — samples has some volatility based on time of day that the sample was taken as well as substances consumed in the prior 48 hours. Remember we are dealing with a lot of fuzzy data — both in what is reported from your sample, and what changes what from the literature.
The result of an uber consensus is a continuation of the prior suggestions (with a little pruning) and incorporation of the current suggestions.
Recent Comments