Foreword – and Reminder
I am not a licensed medical professional and there are strict laws where I live about “appearing to practice medicine”. I am safe when it is “academic models” and I keep to the language of science, especially statistics. I am not safe when the explanations have possible overtones of advising a patient instead of presenting data to be evaluated by a medical professional before implementing.
I cannot tell people what they should take or not take. I can inform people items that have better odds of improving their microbiome as a results on numeric calculations. I am a trained experienced statistician with appropriate degrees and professional memberships.
Backstory of Latest Sample
In light of your recent few blog posts about uploads without many microbiome shifts to work with, I was thinking this could be a beneficial walkthrough video for what seems to be the opposite.
I was doing pretty well on my antibiotic rotations (mainly tetracycline two weeks on, two weeks off since Aug of 2021) until Feb or so when I had a major crash / flare that I’m still suffering from.
I did have a very mild case of Covid in mid January that felt no worse than a regular cold.
But from what little I can parse from this sample, it seems I may be struggling with long Covid. I say little, because my brain fog is extremely dense.
And all of the results I’m getting for this sample via your site seem so drastically different from what has been going on over the last 7 years (my oldest sample is from 2015).
Comparison of samples
This person has samples going back to 2015 using uBiome. Unfortunately for comparison we need to keep to the same lab (why? read The taxonomy nightmare before Christmas…).
Jason Hawrelak Criteria etc
We finally see an improvement with Jason’s criteria. We also may be seeing more diversity with the increase of Genus and Species found. I say may because this could be a side-effect of a low raw count in some samples.
| Date | Percentile | Unhealthy Bacteria | Genus | Species |
| 2022-04-11 | 98.8 %ile | 8 | 220 | 303 |
| 2022-01-11 | 89 %ile | 11 | 89 | 141 |
| 2021-03-09 | 89 %ile | 8 | 108 | 153 |
| 2020-05-27 | 89% ile | 7 | 153 | 223 |
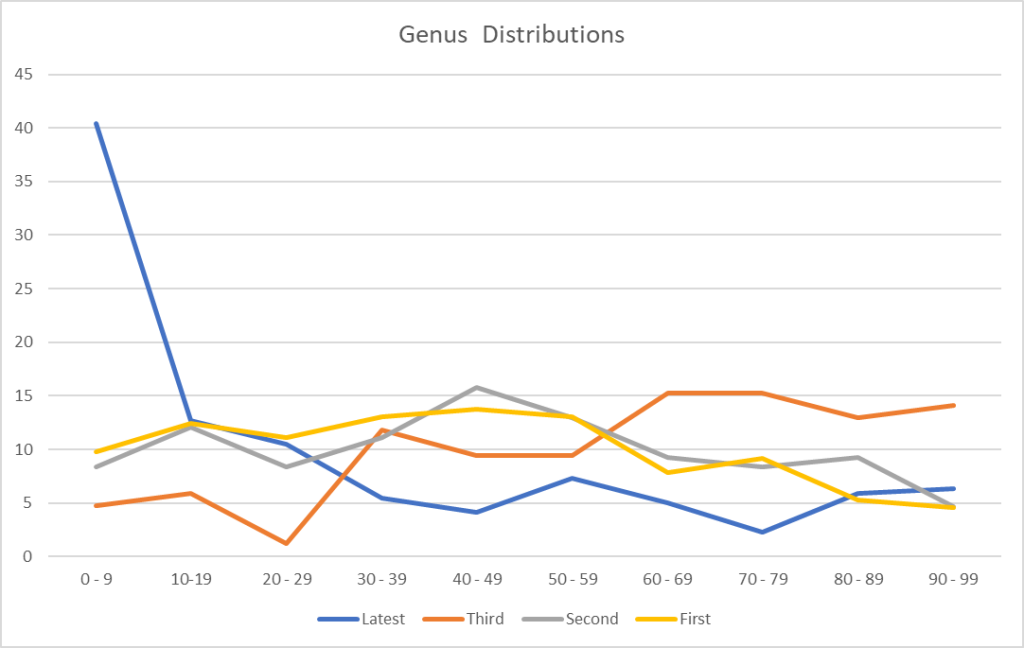
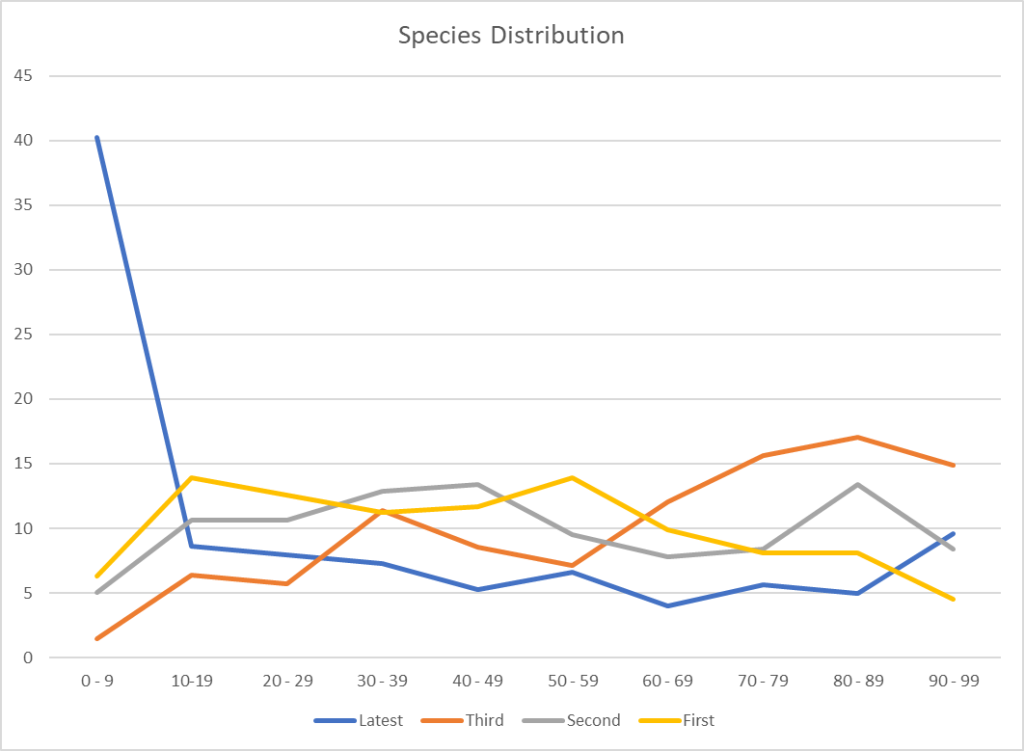
I decided to look at the raw reads (which are captured from Thryve and Biomesights)
| Sample Date | Raw Reads |
| 5/27/2020 | 43311 |
| 3/9/2021 | 29247 |
| 1/11/2022 | 17630 |
| 4/11/2022 | 153194 |
This lead me to look at what typical raw counts are from Ombre/Thryve
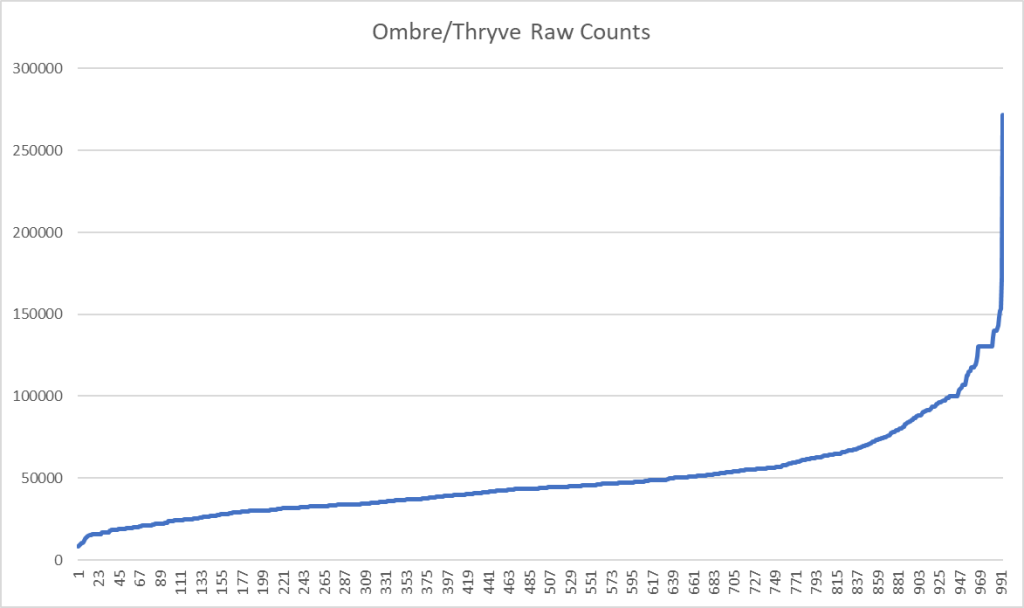
To find the raw counts for your sample, open the csv and look for this line
taxon_id,rank,name,parent,count,...
2,kingdom,Bacteria,,45341,...What is the consequences? It means that rarer bacteria may be ghost-like, appearing or disappearing from sample to sample. This adds let one more layer of fuzziness to doing analysis and generating suggestions.
First Question: ME/CFS or Long COVID microbiome or both?
This person uploaded the Ombre FASTQ files to BiomeSight so I may used data from the Long COVID study there. Both condition present similarly, I am curious to see if we have sufficient reference data to decide which condition is a better match.
| Rank | Name ( 👍 match National Library of Medicine Citations for Long COVID) | Your value | Percentile |
|---|---|---|---|
| clade | FCB group | 25060 | 5.9 |
| class | Bacteroidia 👍 | 22700 | 4.1 |
| class | Betaproteobacteria 👍 | 1510 | 13.3 |
| class | Spirochaetia | 140 | 86.1 |
| family | Bacteroidaceae 👍 | 20690 | 5.4 |
| family | Eubacteriaceae 👍 | 650 | 30.9 |
| genus | Caloramator 👍 [family] | 1520 | 68.5 |
| genus | Nostoc | 20 | 30.6 |
| genus | Roseburia 👍 | 12230 | 34.6 |
| norank | Eubacteriales incertae sedis 👍 [family] | 60 | 11.9 |
| order | Bacteroidales 👍 | 22700 | 4.1 |
| order | Burkholderiales | 1470 | 12.9 |
| phylum | Spirochaetes | 140 | 86 |
| species | Butyrivibrio proteoclasticus 👍[genus] | 10 | 3.6 |
| species | Faecalibacterium prausnitzii 👍 | 301950 | 98.5 |
| species | Roseburia faecis 👍 [family] | 620 | 24.3 |
| Rank | Name (👍 matches National Library of Medicine Citations for Chronic Fatigue Syndrome | Your value | Percentile |
|---|---|---|---|
| family | Halanaerobiaceae | 20 | 37 |
| genus | Anaerovibrio | 570 | 65.1 |
| genus | Finegoldia | 20 | 7 |
| genus | Halanaerobium | 20 | 31.8 |
| genus | Leuconostoc | 10 | 3.2 |
| genus | Pediococcus | 10 | 3.9 |
| order | Syntrophobacterales | 10 | 3.9 |
| species | Anaerotruncus colihominis | 850 | 60.2 |
| species | Anaerovibrio lipolyticus | 570 | 65.4 |
| species | Bacteroides acidifaciens 👎[sibling] | 10 | 0.9 |
| species | Bacteroides fluxus 👎[sibling] | 20 | 9.8 |
| species | Clostridium akagii 👎[sibling] | 10 | 5.5 |
| species | Clostridium cadaveris 👎[sibling] | 10 | 3.8 |
| species | Finegoldia magna | 10 | 1.8 |
| species | Odoribacter denticanis 👍[sibling] | 10 | 2.5 |
| species | Prevotella copri 👍[sibling] | 10 | 0.6 |
We have concurrent matches for both both conditions
- Finegoldia magna, which is not reported in the literature
- The table above hints that he is at present much closer to Long COVID than ME/CFS.
I am not sure about the political correctness of saying “Congrads! You no longer have ME/CFS, you have Long COVID!” is what the microbiome reads like.
What is interesting is that the microbiome constantly shifts/evolves, with Long COVID the infection is constant and the duration since the infection is short — hence less evolution of the microbiome over all patients. With ME/CFS the triggering infection possibilities are huge with 20, 30, 40 years of evolution of the microbiome — hence patterns are diffused by time and original infection.
Looking at deficiency of compounds produced, we see a dramatic drop from the previous sample suggesting that bacteria are getting the needed inputs for correct functioning.
| Sample Date | 1%ile | 5%ile | 10%ile |
| 5/27/2020 | 4 | 14 | 60 |
| 3/9/2021 | 2 | 14 | 16 |
| 1/11/2022 | 197 | 233 | 244 |
| 4/11/2022 | 6 | 28 | 52 |
Where do we go from here
I am going to do consensus, but do only 3 items:
- Hand Picked Bacteria using the study in progress data using BiomeSight (16 bacteria)
- Using US National Library of medicine filter to Long COVID using BiomeSight and Box-Whiskers (14 bacteria)
- Using US National Library of medicine filter to Long COVID using Ombre and Box-Whiskers (14 bacteria)
The consensus is below as a download. Since antibiotics are being prescribed at present, I included that in the suggestions criteria.
Some highlights
- linseed(flaxseed) is #1 by a wide margin
- thiamine hydrochloride (vitamin B1), pyridoxine hydrochloride (vitamin B6), vitamin b3 (niacin), vitamin b7 biotin (supplement) (vitamin B7), Vitamin C (ascorbic acid)
- Cyanocobalamin (Vitamin B-12) and folic acid,(supplement Vitamin B9) has less predicted impact
- B Complex appears negative, as does Vitamin E, D, K2 and A
- glycyrrhizic acid (licorice) – which means to me, usually means Spezzatina
- Then a stack of antibiotics and prescription items (checking items listed on this page)
- I checked the typical ME/CFS ones
- minocycline (antibiotic)s -250 AVOID
- tetracycline (antibiotic)s -207 AVOID
- fluoroquinolone (antibiotic)s -29 AVOID
- rifaximin (antibiotic)s – 134 AVOID
- azithromycin,(antibiotic)s +49
- fludrocortisone acetate,(prescription) +353
- gabapentin,(prescription) +353
- naltrexone hydrochloride dihydrate,(prescription) +353
- nimodipine,(prescription) + 353
- pentoxifylline,(prescription) + 353
- magnesium -180.2 If supplementing, pause
- melatonin supplement + 266.3
- I checked the typical ME/CFS ones
Why did I focus on the ME/CFS ones? Path of least resistance for the prescribing MD – the MD accepts ME/CFS and thus will have low resistance to prescriptions often used for ME/CFS. Asking for them for Long COVID could get rolling of eyes…. As always, we are using these off-label for their computed microbiome effect. For the prescription items, I would suggest rotation (one item for 10 days, then a 0-10 day break, then another item (or repeat if limited to one item).
3 thoughts on “ME/CFS x COVID :- Long COVID instead”
Comments are closed.