This is a thought experiment transformed into an implementation for people to experiment with.
In doing educational reviews of a variety of samples, I came across a person whose progressed had slowed. In trying to understand why [The ME/CFS Quest for Health], I looked at metabolites level between his current sample and previous sample. To my surprise, the highest ones (highest percentile) had barely budgeted.
I looked at the prior Dec 24 sample and compare the KEGG Compounds to the current sample starting with the highest percentile ones:
- All of these were at 99%ile
- 3-Deoxy-D-glycero-D-galacto-non-2-ulopyranosonate – still 99%ile
- Chitobiose – still 99%ile
- Heparan sulfate N-acetyl-alpha-D-glucosaminide – still 99%ile
- D-Cysteine – still 99%ile
- alpha-Kojibiosyldiacylglycerol – still 99%ile
- dTDP-3,4-dioxo-2,6-dideoxy-D-glucose – down to 98%ile
- All of these were at 98%ile
- Urea
- R)-3-(3,4-Dihydroxyphenyl)lactate down to 86%ile
- S-Methyl-5-thio-D-ribose 1-phosphate down to 97%ile
- Coenzyme M 7-mercaptoheptanoylthreonine-phosphate heterodisulfide down to 96%ile
- D-Mannose 1-phosphate down to 96%ile
- 2-Amino-2-deoxy-D-gluconate down to 84%ile
- Retinol down to 96%ile
- (R)-3-(4-Hydroxyphenyl)lactate down to 86%ile
While the bacteria changed, the extreme metabolites remained high but with a few reducing. There is a potential to generate suggestions based on these KEGG compounds — a little messy and definitely pushing inference into new turf.
An Idea
I asked Perplexity.ai on how to reduce a few. A typical response is shown below
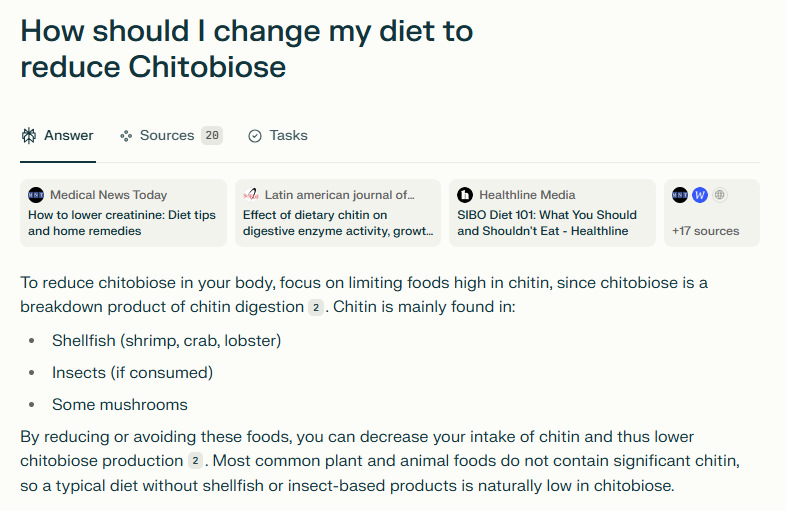
This answer is the typical false logic/inference seen with “To reduce cholesterol, just eat food low in cholesterol“.
Possible Expert System Algorithm
On MicrobiomePrescription.com, the suggestion algorithm works solely off the bacteria that is reported by the microbiome test. This is done by using facts harvested from US National Library of Medicine studies. There are no (or likely extremely few) studies dealing with diet and metabolites.
The key phrase is reported by. We know that reporting is not standardized and often using only 16s.
Idea!
Current logic on MicrobiomePrescription.com is bacteria => suggestion impact. What if we add another approach: metabolite => normalized bacteria distribution => suggestions. We want this to have less randomness than 16s. The folks at PrecisionBiome.Eu shared 1000 shotgun results from healthy individuals with me so I could construct a normalized bacteria distribution model. From this model, I computed metabolites using data from KEGG: Kyoto Encyclopedia of Genes and Genomes and ended up with a facts table consisting of:
- Metabolite
- Suggestion / Modifier
- Estimated Impact
The metabolite is identified by KEGG ID.
Implementation
Since the microbiome and its metabolites are very interconnected and interact with each other. I decided that looking at the top and bottom 5-10%ile (i.e. those with a percentile ranking of 90-95%ile or higher, a percentile of 10-5%ile or lower) was a reasonable approach. There is a little trust that the central limit theorem will generate reasonable results and allow metagenomics to be directly used for getting suggestions.
On the [Research Features] tab, this panel has been added:
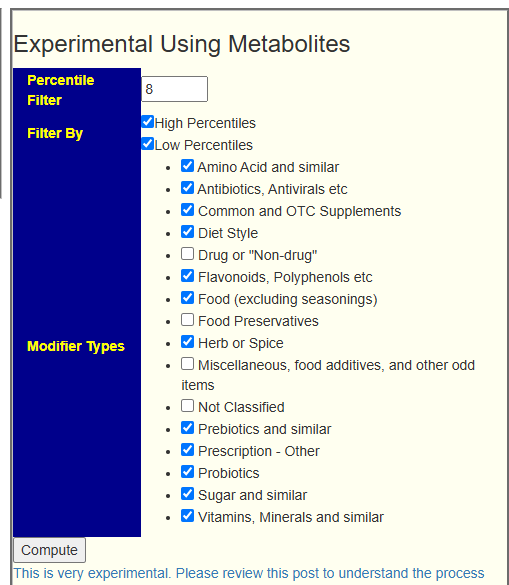
This produces a report listing the Metabolites targeted (High and/or Low) and then Suggestions


Observation
To me, what I found very interesting is that there are a few that are very high in impact with rapid drop off. This means there are only a few critical items to add to the general bacteria-based suggestions.
Recent Comments