For almost a decade I have suspected that there was an interaction between the microbiome and Antiphospholipid syndrome (APS) also known as Hughes Syndrome (after the MD, see below). This is also called “sticky blood syndrome” [HealthLine]. For some researchers, it is deemed to be a significant contributor to fatigue in Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) [1999 D. Berg] and likely also applies to Long COVID. My own singleton experience seems to confirm it for myself.
A reader asked about phospholipids on Facebook today, so I revisited available literature
This article by Graham R.V. Hughes, MD, FRCP (the discoverer) in 2016 is well worth reading.
For me, APS/Hughes syndrome is very much a neurological condition. Brain function does seem to be especially targeted—the more APS patients one sees, the wider and wider the neuropsychiatric ripples spread.
APS: What Rheumatologists Should Know about Hughes Syndrome • By Graham R.V. Hughes, MD, FRCP
Of course, running off the experience of just one, or even a few people, is not the best practice. Testimonials suck because of rose color glasses, fake testimonials, mainly positive responders report, and placebo effects. So what does the literature state. First there is some literature that are general discussions without the type of detail that I would love to see:
- Crosstalk Between the Gut Microbiome and Bioactive Lipids: Therapeutic Targets in Cognitive Frailty [2020]
- Phospholipid catabolism by gut microbiota and the risk of cardiovascular disease [2013]
Then we come to this article: Phosphatidylglycerols are induced by gut dysbiosis and inflammation, and favorably modulate adipose tissue remodeling in obesity [2019] which uses one of my favorite information source, the Kyoto Encyclopedia of Genes and Genomes. “We found that PGs were positively associated with microbiomes enriched with endotoxin-synthesis genes and associated with markers of inflammation.”
- ” In antiphospholipid syndrome, a thrombotic auto-immune disorder, autoreactive T cells and antibodies cross-react with auto-antigen mimicking peptides from gut commensals which appears to contribute to the pathophysiology. ” Gut microbiota and their metabolites in cardiovascular disease, 2021
Digging further we find:
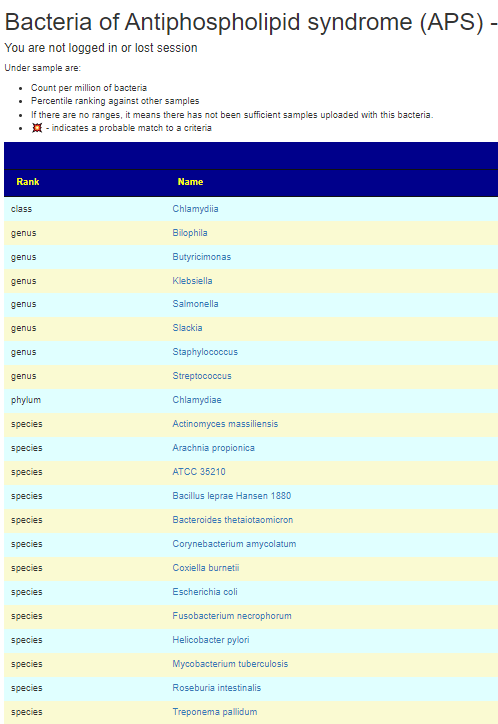
Bacteroides thetaiotaomicron, Actinomyces massiliensis, Pseudopropionibacterium propionicum, Corynebacterium amycolatum, Ruminococcus gnavus and Roseburia intestinalis[2021] lead to the formation of pathogenic T‑cell and autoantibody responses via the cross-reactivity with autoantigens (Ro60, dsDNA and ß2 glycoprotein I).
The role of the microbiome in lupus and antiphospholipid syndrome [2020]
M. pneumoniae and Streptococcus spp. infections, which are among the most prevalent bacterial infections in children and young adults, were linked to the occurrence of aPL. …. an anaerobic bacterium Fusobacterium necrophorum, although a variety of other bacteria such as streptococci, staphylococci, and enterococci may be also responsible…. a specific change in the gut microbial composition in APS patients. Particularly, a decrease of bacteria belonging to the genus Bilophila and overgrowth of bacteria of the Slackia genus were shown… enrichment by Slackia spp. and by the lower abundance of butyrate-producing Butyricimonas
Environmental Triggers of Autoreactive Responses: Induction of Antiphospholipid Antibody Formation [2019]
More discussion of mechanism is in The Role of the Gut Microbiota in the Pathogenesis of Antiphospholipid Syndrome [2015]
Bottom Line
APS only requires one of the bacteria above to trigger it. In terms of using Microbiome Prescription, I would look at Bilophila and Butyricimonas – if below 50%ile, hand pick it, then look at Slackia, if above 50%ile then hand pick it. Check the other bacteria cited above, and if any are over 75%ile, hand pick those. “It only takes one rotten apple to spoil the barrel” seems to apply here.
I have added APS to my PubMed reference list:
Personal Observations
I checked my samples from my last ME/CFS flare and found that Bacteroides thetaiotaomicron went from 73%ile on first sample after onset, to 96%ile on second sample, down to 79%ile, then 70%ile then 20%ile a few months later with recovery and returning to work. The key triggering bacteria will likely be different for each person but you at least have a candidate list to work from.
Recent Comments