See this post for the criteria being used.
| Accessibility: A- | CSV and FastQ (by special request, not on website) |
| Significance: C- | Ranges for a few taxa only |
| Reference: D | Diet styles could be selected for actions |
| Actions: B | 634 items estimated (only items that increase a taxa) |
| Evidence: A | References provided |
| Benevolence: D | Sells their own probiotics |
| Support: D | Multiple reports of incorrect shipments, slow response |
| Promptness: C | 2-3 weeks |
| Metadata: D | Nothing |
| Overtime: D | Not provided |
| Taxa Score: 40% – B | Less in common with uBiome |
| Taxa Scope: 111% | More Reported |
Actions are for only increasing a small number of Taxa
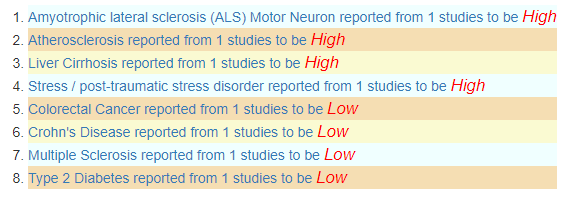
Note: Many of these bacteria with too high a level are associated with medical conditions.
Analysis
- Wellness score above their average – no information on percentile rank
- Diversity score above their average
- They use healthy population from the American Gut Project. American Gut only does a few of the bacteria reported by Thryve. Data does not really apply.
When I processed it thru Jason Hawrelak guidelines, more than 50% are of concern and some are overgrowths.
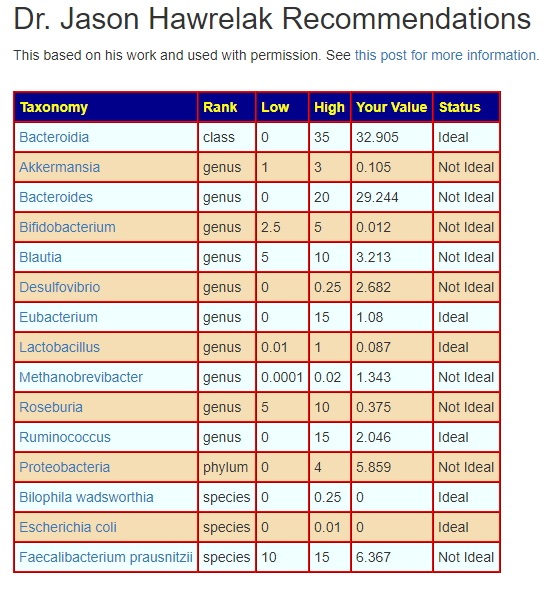
Two Different Scalings?
The site leaves the user confused because it gives two different percentages for many items as shown below.
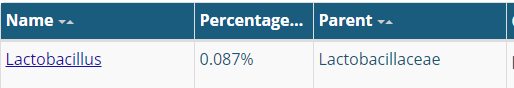
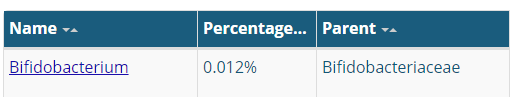
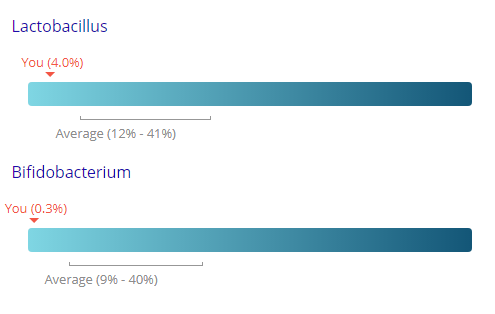
In discussion, it was suggested that this may be the percentage of the ranges of values that they have seen. Assuming this is the case, we can compute the actual ranges (per Jason numbers)
- Lactobacillus: 0.261% – 0.88% (range is inside of Jason’s range)
- Bifidobacterium: 0.9% – 1.33% (range is entirely below Jason’s range)
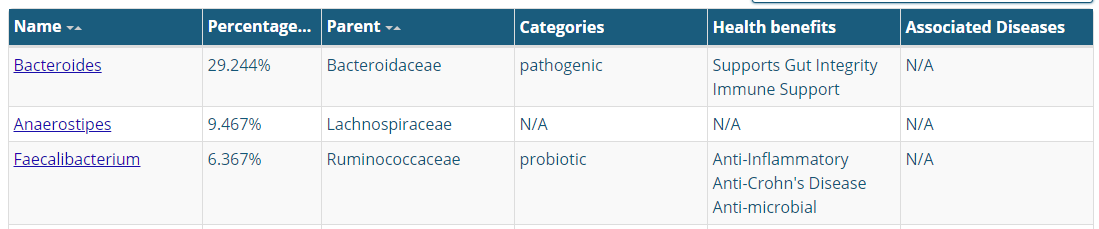
Medical Material Provided
No ranges (or even averages)


Web Site is Sluggish
Lots of spinning devices that often went on for minutes.
Bottom Line
Some well documented actions. For most taxa, no reference range or even average from their samples are given. Test results are usable for processing elsewhere. They document the strain of probiotics they suggest and provide explanations.
They have some good strengths, but also weakness which I hope they will address.
Recent Comments