This set of tools is designed for two scenarios:
- One person who has had many samples of time. Typically that person is looking for outliers to reduce or disappear
- A family with samples from different members. Typically one person is challenged and since the family group has shared DNA and diet — the hope is that the bacteria grouping causing the challenge will be identified. Once identified, it may be actionable.
- This scenario will have more tools added over the next weeks.
I have used them in these prior posts:
If you have two or more samples uploaded, you will see the top two items on the Available Samples page. These may be collapsed into one over the next few weeks.
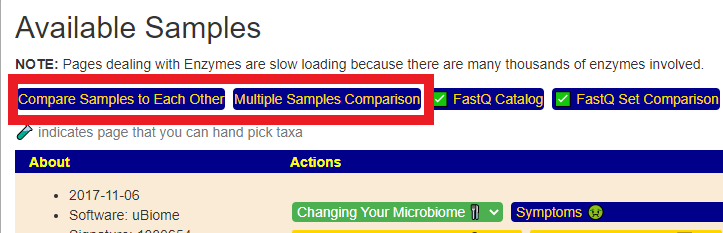
Clicking the right button of these two will take you to a sample selection page.
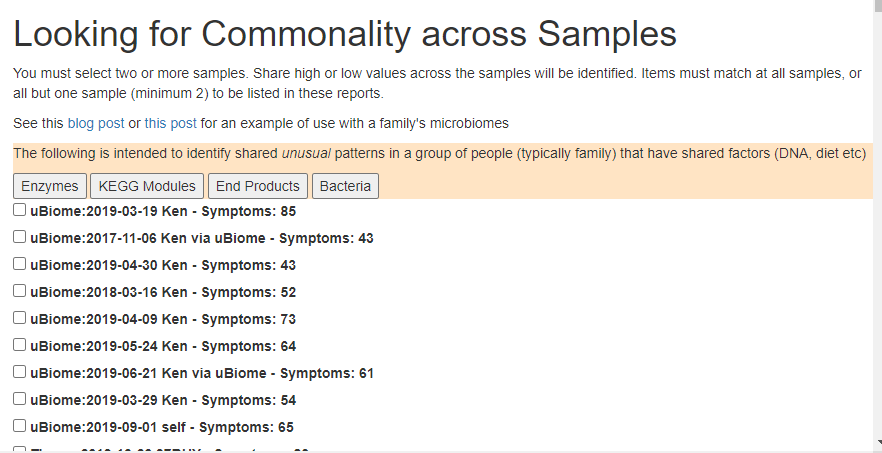
The program will list all items below that matches all samples OR all samples except 1 (but at least two).
I selected a group of 5 samples from when I was having a ME/CFS flare.
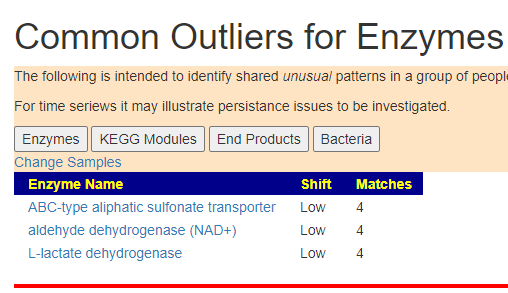
IMHO, it correctly identified what was wrong with me.
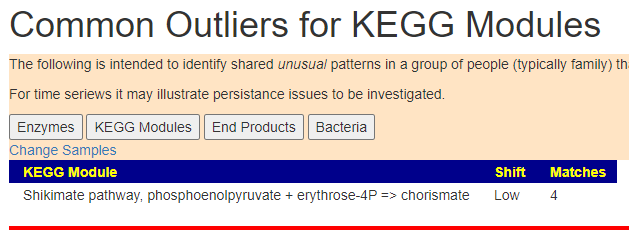
Doing some research, I found “. L-tryptophan is produced in the shikimate pathway from chorismate” Which lead to many ME/CFS articles on PubMed.
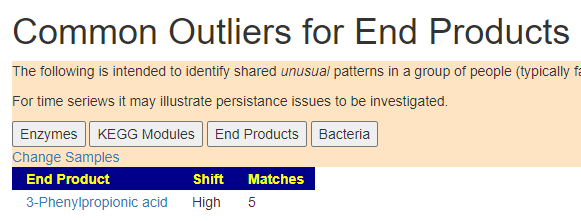
Again, it may take some research to understand what this is.
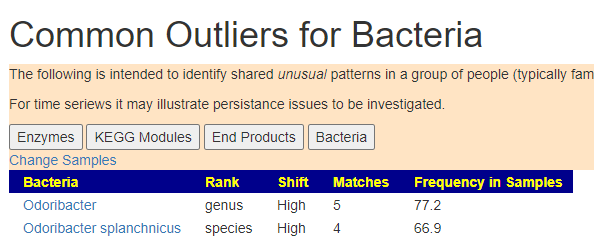
The cause of ME/CFS flare was stress and a quick search on PubMed found several articles where this genus increased with stress, the latest was “Gut Microbiota Are Associated With Psychological Stress-Induced Defections in Intestinal and Blood–Brain Barriers“[2020]
Bottom Line
This set of tools does not give immediate answers; it gives you leads to investigate. For myself, the findings plus the use of PubMed studies weaved a story of what happened that agrees with the literature. This is very important because ME/CFS contains dozens of subsets. Often I have seen that what is helpful for one subset is harmful for another. I suspect this also applies to other conditions, such as ASD/Autism.
In this case, it identified one key family to reduce. Identified enzymes that I was short on. Lead to a possible supplements that I should consider because of the dysruption.
Recent Comments