Part of being open sources is being open-source on where you are getting information. For myself, my primary criteria is published peer-reviewed scientific papers. You must determine your own criteria (for example, using unpublished studies referenced and summarized in patent filings).
Citations are the connectors between bacteria, probiotics, medical conditions, lab measurements etc. They are the evidence chain of how things are related.
In this post I will describe column by column what I feel should be in your citation table as a minimum:
- CitationId – unique identifier used in relationships : int
- Title – The title of the study or document: nvarchar(255)
- CitationJson – A summary using the PubMed pattern: nvarchar(max)
- RawText – A pure text version of the document. It will be used for Text Mining and manual inspection : nvarchar(max)
- RawDocument – A binary version of the document. Often this will be a PDF file or may be an image. varbinary(max)
- RawDocumentMimeType – The mime type for the RawDocument: varchar(60)
- Summary – This is typically the PubMed summary for the study: nvarchar(max)
- PMID: PubMed ID : int
- PMCID: PubMed Citation Id: varchar(14)
- DOI: Digital Object Identifier (DOI) : varchar(60)
Ideally, a component should extract the RawDocument into RawText. For example, OCR for images and various converters for PDF. This type of conversion is outside of the scope of this blog series. They can be done (I have written such professionally).
Source: https://github.com/Lassesen/Microbiome2/tree/master/CitationsConsole
Below are the class diagrams:
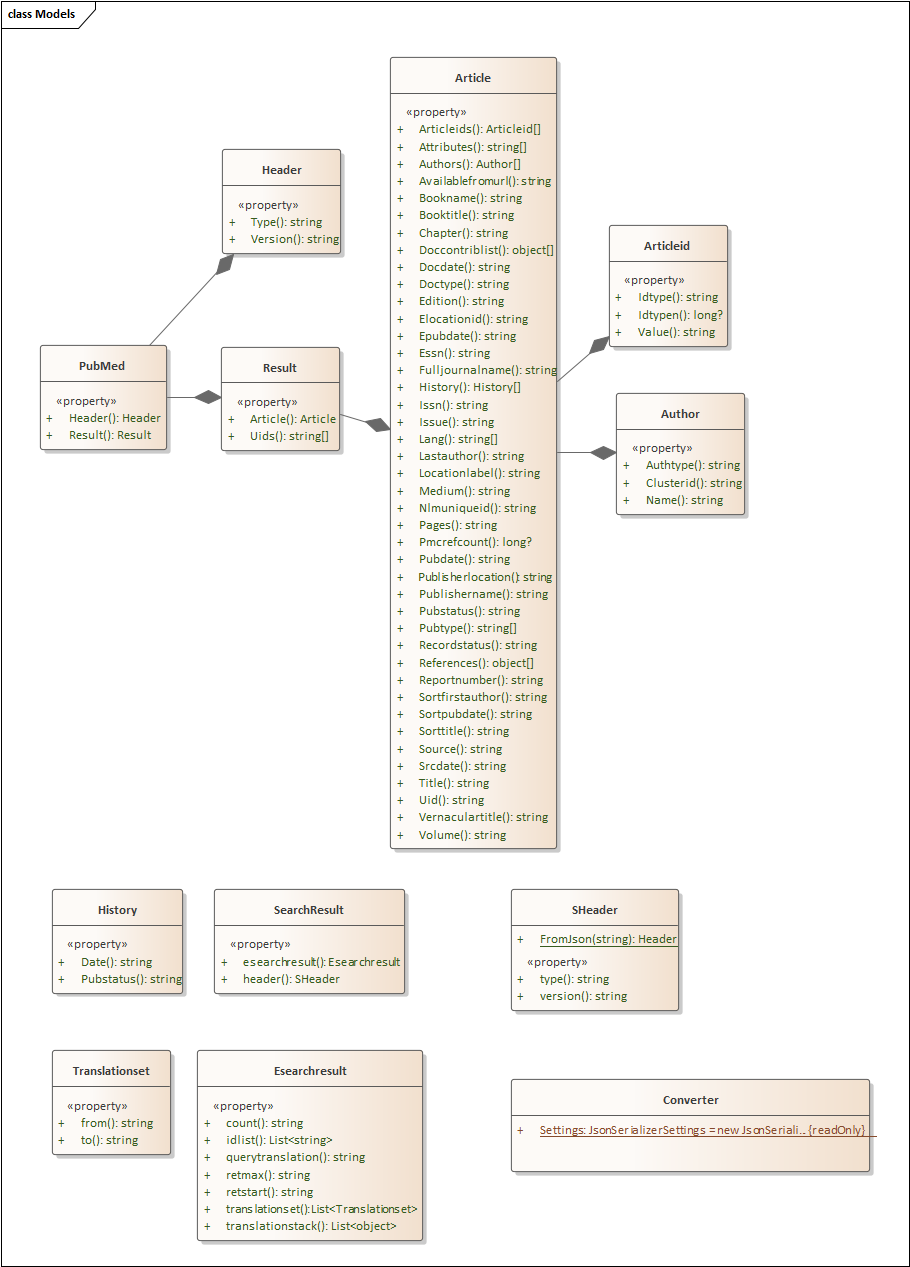
For some background on the PubMed API calls, see this page. The
The following shows the response searching for “Corgi”.
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=corgi&retmode=json&retmax=10
{
"header": {
"type": "esearch",
"version": "0.3"
},
"esearchresult": {
"count": "101",
"retmax": "10",
"retstart": "0",
"idlist": [
"31194653",
"31172540",
"30577744",
"29794371",
"29681557",
"29355995",
"28942296",
"28933509",
"28855429",
"28566164"
],
"translationset": [
],
"translationstack": [
{
"term": "corgi[All Fields]",
"field": "All Fields",
"count": "101",
"explode": "N"
},
"GROUP"
],
"querytranslation": "corgi[All Fields]"
}
}
Running it is simple, just give it the name of the condition (it defaults to 100 citations, but you can increase it. I usually run with 10,000 citations at a time for each bacteria taxonomy name).
CitationConsole “Some Unusual Condition”
What is the net result? Running it with “SIBO”, we see in the database.
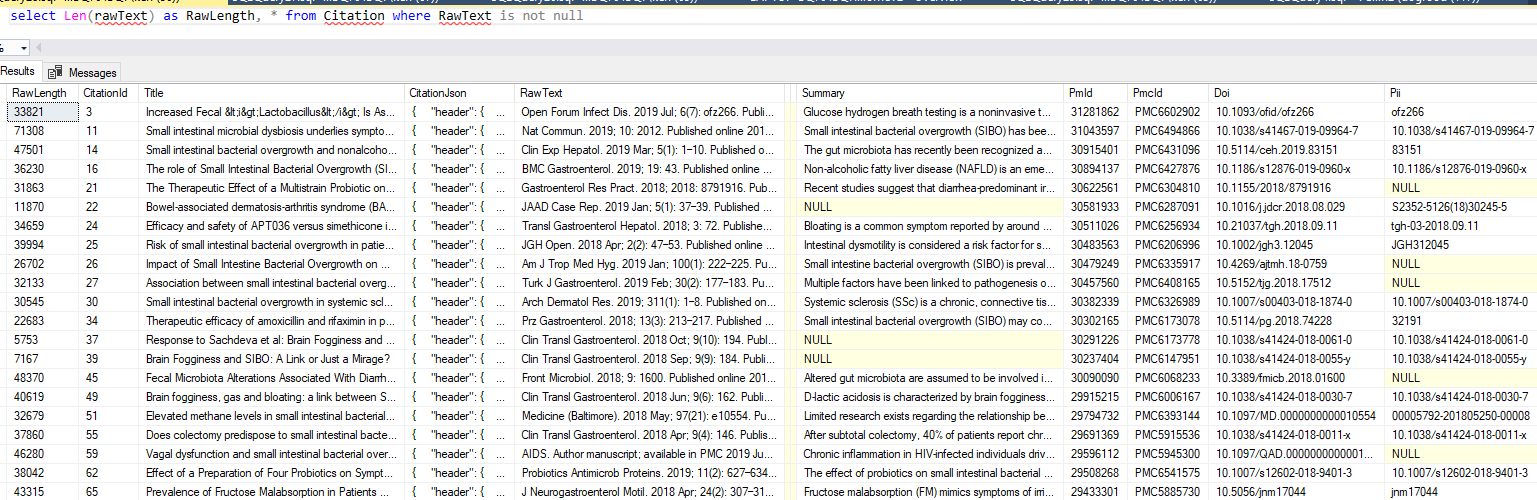
So we have our data ready for either text mining or manual review.
Home Work
We can now fill up our citation tables, in some cases with the complete text (excluding appendices) as well as a short summary. How are you going to process the data?
- Manual review?
- Use Text Data Mining? See this page to start.
Processing this data is where there is a major effort needed.
Recent Comments