A post on Facebook reminded me of an update from a recent data addition. We have added the prevalence of each condition. If you are at 75% ile for a condition that impacts only 5% of the population — there is low risk. But if the conditions impacts 50% of the population, you are at a significant risk.
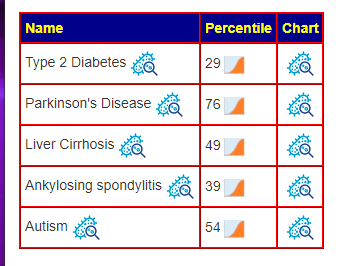
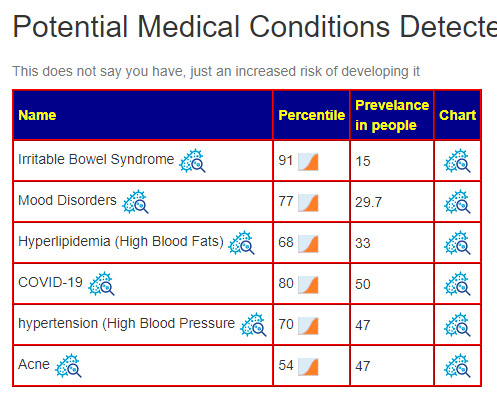
Actions?
First, you should inform your medical professional that you are concerned about elevated risk for conditions that you do not have and inquire about any testing. Do not be surprised at some “rolling of the eyes” on how this risk was determined. Remember — many of the drugs prescribed for conditions do alter your microbiome in a favorable way (academic question: is this a possible mechanism of action?)
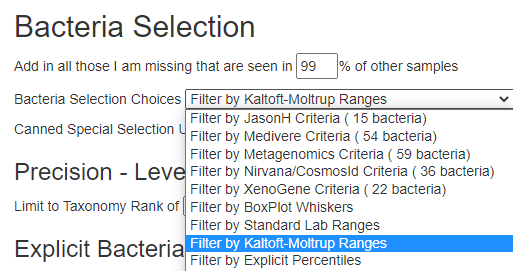
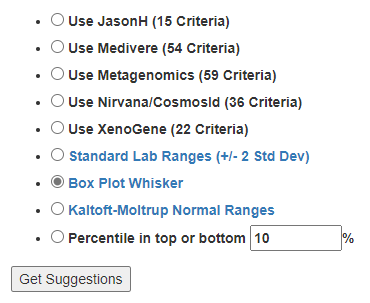
Second, for items of the greatest concern, you may wish filter by the bacteria associated with this condition to get suggestions that may shift your microbiome away from this profile.
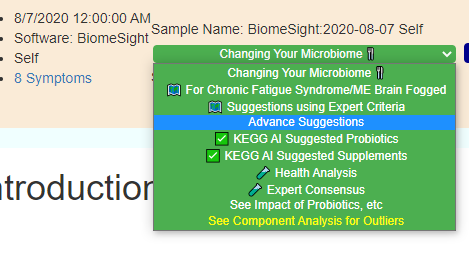
Click on that link and then
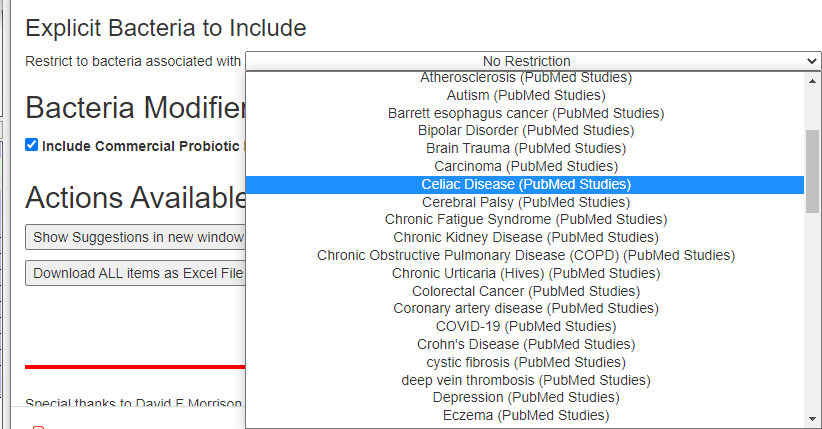
You may wish to play around with different filtering criteria


Recent Comments