| Enzyme Name | ECKEY | With
Long COVID | Without
Long
COVID | T-Score | DF | Probability |
| (S)-3-hydroxy-3-methylglutaryl-CoA acetoacetate-lyase (acetyl-CoA-forming) | 4.1.3.4 | 130357 | 52076 | 13.57 | 666 | P < 0.001 |
| 4-amino-5-aminomethyl-2-methylpyrimidine aminohydrolase | 3.5.99.2 | 151693 | 69126 | 13.48 | 666 | P < 0.001 |
| dihydrourocanate:acceptor oxidoreductase | 1.3.99.33 | 135133 | 55921 | 13.44 | 666 | P < 0.001 |
| nucleoside-triphosphate diphosphohydrolase | 3.6.1.9 | 145528 | 63523 | 13.38 | 666 | P < 0.001 |
| poly(deoxyribonucleotide)-3′-hydroxyl:5′-phospho-poly(deoxyribonucleotide) ligase (ATP or NAD+) | 6.5.1.6 | 129475 | 51779 | 13.34 | 666 | P < 0.001 |
| poly(deoxyribonucleotide)-3′-hydroxyl:5′-phospho-poly(deoxyribonucleotide) ligase (ATP, ADP or GTP) | 6.5.1.7 | 129475 | 51779 | 13.34 | 666 | P < 0.001 |
| acetyl-CoA:kanamycin-B N6′-acetyltransferase | 2.3.1.82 | 128673 | 51662 | 13.32 | 666 | P < 0.001 |
| acetyl-CoA:2-deoxystreptamine-antibiotic N3-acetyltransferase | 2.3.1.81 | 130200 | 53104 | 13.28 | 666 | P < 0.001 |
| (1->4)-alpha-D-galacturonan reducing-end-disaccharide-lyase | 4.2.2.9 | 128038 | 50989 | 13.28 | 666 | P < 0.001 |
| alpha-maltose-6′-phosphate 6-phosphoglucohydrolase | 3.2.1.122 | 130496 | 53951 | 13.28 | 666 | P < 0.001 |
| D-serine ammonia-lyase (pyruvate-forming) | 4.3.1.18 | 128576 | 51987 | 13.23 | 666 | P < 0.001 |
| ATP phosphohydrolase (ABC-type, iron(III) enterobactin-importing) | 7.2.2.17 | 130373 | 54180 | 13.15 | 666 | P < 0.001 |
| protein-Npi-phospho-L-histidine:D-mannose Npi-phosphotransferase | 2.7.1.191 | 140701 | 62807 | 13.14 | 666 | P < 0.001 |
| protein-Npi-phospho-L-histidine:D-mannitol Npi-phosphotransferase | 2.7.1.197 | 129284 | 53723 | 13.06 | 666 | P < 0.001 |
| ADP-alpha-D-glucose:alpha-D-glucose-1-phosphate 4-alpha-D-glucosyltransferase (configuration-retaining) | 2.4.1.342 | 134654 | 58028 | 13.04 | 665 | P < 0.001 |
| aryl-ester hydrolase | 3.1.1.2 | 151171 | 72377 | 13.02 | 666 | P < 0.001 |
| ATP phosphohydrolase (ABC-type, Fe3+-transporting) | 7.2.2.7 | 142152 | 66111 | 12.86 | 666 | P < 0.001 |
| palmitoyl-CoA hydrolase | 3.1.2.2 | 149499 | 71728 | 12.83 | 666 | P < 0.001 |
| ATP:[protein]-L-tyrosine O-phosphotransferase (non-specific) | 2.7.10.2 | 131925 | 57360 | 12.75 | 666 | P < 0.001 |
| D-psicose 3-epimerase | 5.1.3.30 | 143416 | 68284 | 12.74 | 666 | P < 0.001 |
| D-tagatose 3-epimerase | 5.1.3.31 | 143416 | 68284 | 12.74 | 666 | P < 0.001 |
| penicillin amidohydrolase | 3.5.1.11 | 139138 | 66165 | 12.62 | 666 | P < 0.001 |
| D-aspartate:[beta-GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)]n ligase (ADP-forming) | 6.3.1.12 | 145356 | 70261 | 12.57 | 666 | P < 0.001 |
| 2′-(5-triphosphoribosyl)-3′-dephospho-CoA:apo-[citrate (pro-3S)-lyase] 2′-(5-phosphoribosyl)-3′-dephospho-CoA-transferase | 2.7.7.61 | 149196 | 74697 | 12.46 | 666 | P < 0.001 |
| ATP:3′-dephospho-CoA 5-triphospho-alpha-D-ribosyltransferase | 2.4.2.52 | 149957 | 75781 | 12.39 | 666 | P < 0.001 |
| 2,4,6/3,5-pentahydroxycyclohexanone 2-isomerase | 5.3.99.11 | 146254 | 73538 | 12.37 | 666 | P < 0.001 |
| acetyl-CoA:citrate CoA-transferase | 2.8.3.10 | 150367 | 76318 | 12.36 | 666 | P < 0.001 |
| L-aspartate:tRNAAsx ligase (AMP-forming) | 6.1.1.23 | 132939 | 61720 | 12.22 | 666 | P < 0.001 |
| 4-phospho-D-erythronate:NAD+ 3-oxidoreductase | 1.1.1.409 | 134073 | 62914 | 12.19 | 666 | P < 0.001 |
| 4-phospho-D-threonate:NAD+ 3-oxidoreductase | 1.1.1.408 | 134073 | 62914 | 12.19 | 666 | P < 0.001 |
| poly(deoxyribonucleotide)-3′-hydroxyl:5′-phospho-poly(deoxyribonucleotide) ligase (ATP) | 6.5.1.1 | 143883 | 68128 | 12.15 | 666 | P < 0.001 |
| ATP:D-erythronate 4-phosphotransferase | 2.7.1.220 | 133631 | 62696 | 12.13 | 666 | P < 0.001 |
| ATP:D-threonate 4-phosphotransferase | 2.7.1.219 | 133631 | 62696 | 12.13 | 666 | P < 0.001 |
| ATP:D-glycero-alpha-D-manno-heptose 7-phosphate 1-phosphotransferase | 2.7.1.168 | 156157 | 78491 | 11.99 | 666 | P < 0.001 |
| 2-lysophosphatidylcholine acylhydrolase | 3.1.1.5 | 160874 | 86814 | 11.71 | 666 | P < 0.001 |
| UDP-alpha-D-glucose:1,2-diacyl-sn-glycerol 3-alpha-D-glucosyltransferase | 2.4.1.337 | 159960 | 87248 | 11.57 | 666 | P < 0.001 |
| N-succinyl-LL-2,6-diaminoheptanedioate amidohydrolase | 3.5.1.18 | 151482 | 82849 | 11.49 | 666 | P < 0.001 |
| acetate:holo-[citrate-(pro-3S)-lyase] ligase (AMP-forming) | 6.2.1.22 | 163245 | 90493 | 11.44 | 666 | P < 0.001 |
| D-tagatose 1,6-bisphosphate D-glyceraldehyde-3-phosphate-lyase (glycerone-phosphate-forming) | 4.1.2.40 | 140814 | 72879 | 11.44 | 666 | P < 0.001 |
| L-iditol:NAD+ 2-oxidoreductase | 1.1.1.14 | 181524 | 109886 | 11.34 | 666 | P < 0.001 |
| alkylated-DNA glycohydrolase (releasing methyladenine and methylguanine) | 3.2.2.21 | 172169 | 89028 | 11.33 | 666 | P < 0.001 |
| oligosaccharide 6-alpha-glucohydrolase | 3.2.1.10 | 161628 | 93712 | 11.12 | 666 | P < 0.001 |
| (3S)-citryl-CoA oxaloacetate-lyase (acetyl-CoA-forming) | 4.1.3.34 | 185375 | 105009 | 10.73 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:tRNA (adenine22-N1)-methyltransferase | 2.1.1.217 | 157938 | 92817 | 10.69 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:16S rRNA (cytidine1409-2′-O)-methyltransferase | 2.1.1.227 | 183689 | 114027 | 10.56 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:23S rRNA (cytidine1920-2′-O)-methyltransferase | 2.1.1.226 | 183689 | 114027 | 10.56 | 666 | P < 0.001 |
| sn-glycerol 3-phosphate:quinone oxidoreductase | 1.1.5.3 | 177064 | 110486 | 10.54 | 666 | P < 0.001 |
| L-glutamate:tRNAGlx ligase (AMP-forming) | 6.1.1.24 | 165448 | 97402 | 10.38 | 666 | P < 0.001 |
| D-glycero-D-manno-heptose 7-phosphate aldose-ketose-isomerase | 5.3.1.28 | 213475 | 135248 | 10.25 | 666 | P < 0.001 |
| beta-D-glucose 1,6-phosphomutase | 5.4.2.6 | 253383 | 169933 | 8.98 | 666 | P < 0.001 |
| type II site-specific deoxyribonuclease | 3.1.21.4 | 245864 | 165266 | 8.77 | 666 | P < 0.001 |
| uroporphyrinogen-III carboxy-lyase (coproporphyrinogen-III-forming) | 4.1.1.37 | 266562 | 184417 | 8.59 | 666 | P < 0.001 |
| ATP phosphohydrolase (ABC-type, Ni2+-importing) | 7.2.2.11 | 141054 | 86367 | 8.56 | 666 | P < 0.001 |
| acetyl-CoA:oxalate CoA-transferase | 2.8.3.19 | 133160 | 80149 | 8.41 | 666 | P < 0.001 |
| 5-phospho-alpha-D-ribose 1,2-cyclic phosphate 1,2-diphosphophosphohydrolase | 3.1.4.57 | 134813 | 82183 | 8.33 | 666 | P < 0.001 |
| Carboxypeptidase Taq | 3.4.17.19 | 136153 | 84074 | 8.31 | 666 | P < 0.001 |
| (2E)-2-enoyl-CoA:NADP+ 4-oxidoreductase | 1.3.1.34 | 134146 | 81935 | 8.31 | 666 | P < 0.001 |
| 2′-deoxyribonucleoside 5′-monophosphate phosphohydrolase | 3.1.3.89 | 136444 | 84251 | 8.19 | 666 | P < 0.001 |
| cellobiose 2-epimerase | 5.1.3.11 | 224933 | 147685 | 8.17 | 666 | P < 0.001 |
| type III site-specific deoxyribonuclease | 3.1.21.5 | 200155 | 141657 | 8.10 | 666 | P < 0.001 |
| D-galactonate hydro-lyase (2-dehydro-3-deoxy-D-galactonate-forming) | 4.2.1.6 | 138933 | 88181 | 8.02 | 666 | P < 0.001 |
| protein-Npi-phospho-L-histidine:maltose Npi-phosphotransferase | 2.7.1.208 | 138609 | 88325 | 7.98 | 666 | P < 0.001 |
| L-aspartate:NAD(P)+ oxidoreductase (deaminating) | 1.4.1.21 | 245150 | 169471 | 7.90 | 666 | P < 0.001 |
| protein-Npi-phospho-L-histidine:sucrose Npi-phosphotransferase | 2.7.1.211 | 144741 | 96084 | 7.85 | 666 | P < 0.001 |
| protein-Npi-phospho-L-histidine:N-acetyl-D-glucosamine Npi-phosphotransferase | 2.7.1.193 | 152088 | 103970 | 7.64 | 666 | P < 0.001 |
| N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol 4-epimerase | 5.1.3.26 | 259434 | 182387 | 7.64 | 666 | P < 0.001 |
| UDP-sugar sugarphosphohydrolase | 3.6.1.45 | 156303 | 108944 | 7.63 | 666 | P < 0.001 |
| rubredoxin:superoxide oxidoreductase | 1.15.1.2 | 147572 | 101699 | 7.40 | 666 | P < 0.001 |
| UDP-2-acetamido-2,6-dideoxy-L-talose:NADP+ oxidoreductase | 1.1.1.367 | 244790 | 176296 | 7.33 | 666 | P < 0.001 |
| (4-O-methyl)-D-glucuronate—lignin ester hydrolase | 3.1.1.117 | 265763 | 190685 | 7.31 | 666 | P < 0.001 |
| alkylated-DNA glycohydrolase (releasing methyladenine and methylguanine) | 3.2.2.20 | 306951 | 231407 | 7.30 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:16S rRNA (cytosine967-C5)-methyltransferase | 2.1.1.176 | 192348 | 143978 | 7.28 | 666 | P < 0.001 |
| ATP:molybdopterin-synthase adenylyltransferase | 2.7.7.80 | 297993 | 218788 | 7.25 | 666 | P < 0.001 |
| UDP-N-acetyl-alpha-D-glucosamine hydro-lyase (inverting; UDP-2-acetamido-2,6-dideoxy-beta-L-arabino-hex-4-ulose-forming) | 4.2.1.115 | 287717 | 211984 | 7.24 | 666 | P < 0.001 |
| UDP-2-acetamido-2,6-dideoxy-beta-L-talose 2-epimerase | 5.1.3.28 | 245857 | 178308 | 7.23 | 666 | P < 0.001 |
| ATP phosphohydrolase (ABC-type, molybdate-importing) | 7.3.2.5 | 157815 | 111902 | 7.18 | 666 | P < 0.001 |
| sucrose:(1->4)-alpha-D-glucan 4-alpha-D-glucosyltransferase | 2.4.1.4 | 148095 | 102284 | 7.16 | 666 | P < 0.001 |
| UDP-N-acetyl-2-amino-2-deoxy-alpha-D-glucuronate:NAD+ 3-oxidoreductase | 1.1.1.335 | 172209 | 124841 | 7.08 | 666 | P < 0.001 |
| an acyl-[acyl-carrier protein]:phosphate acyltransferase | 2.3.1.274 | 185445 | 139090 | 7.05 | 666 | P < 0.001 |
| FMNH2:NAD(P)+ oxidoreductase | 1.5.1.39 | 156478 | 107179 | 7.02 | 666 | P < 0.001 |
| cellobiose:phosphate alpha-D-glucosyltransferase | 2.4.1.20 | 165795 | 117785 | 6.98 | 666 | P < 0.001 |
| 4-beta-D-xylan xylohydrolase | 3.2.1.37 | 308412 | 233667 | 6.87 | 666 | P < 0.001 |
| crossover junction endodeoxyribonuclease | 3.1.21.10 | 341116 | 260719 | 6.82 | 666 | P < 0.001 |
| nucleoside-2′,3′-cyclic-phosphate 3′-nucleotidohydrolase | 3.1.4.16 | 321464 | 242830 | 6.82 | 666 | P < 0.001 |
| UDP-glucuronate 4-epimerase | 5.1.3.6 | 280650 | 207501 | 6.80 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:precorrin-2 C20-methyltransferase | 2.1.1.130 | 293271 | 227548 | 6.79 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:cobalt-factor-II C20-methyltransferase | 2.1.1.151 | 293124 | 227408 | 6.78 | 666 | P < 0.001 |
| dipeptidase E | 3.4.13.21 | 204290 | 149503 | 6.73 | 666 | P < 0.001 |
| GTP:alpha-D-mannose-1-phosphate guanylyltransferase | 2.7.7.13 | 331462 | 252998 | 6.69 | 666 | P < 0.001 |
| ATP:1,2-diacyl-sn-glycerol 3-phosphotransferase | 2.7.1.107 | 333291 | 259697 | 6.61 | 666 | P < 0.001 |
| L-selenocysteine selenide-lyase (L-alanine-forming) | 4.4.1.16 | 337172 | 260622 | 6.56 | 666 | P < 0.001 |
| 1,4-beta-D-mannooligosaccharide:phosphate alpha-D-mannosyltransferase | 2.4.1.319 | 300427 | 227352 | 6.54 | 666 | P < 0.001 |
| 4-O-beta-D-mannopyranosyl-N-acetyl-D-glucosamine:phosphate alpha-D-mannosyltransferase | 2.4.1.320 | 300427 | 227352 | 6.54 | 666 | P < 0.001 |
| sortase B | 3.4.22.71 | 161358 | 119363 | 6.52 | 666 | P < 0.001 |
| cobalt-precorrin 5A acylhydrolase | 3.7.1.12 | 160731 | 118900 | 6.48 | 666 | P < 0.001 |
| 6-carboxy-5,6,7,8-tetrahydropterin ammonia-lyase | 4.3.99.3 | 329010 | 253328 | 6.47 | 666 | P < 0.001 |
| precorrin-8 11,12-methylmutase | 5.4.99.60 | 161359 | 119806 | 6.46 | 666 | P < 0.001 |
| precorrin-8X 11,12-methylmutase | 5.4.99.61 | 161359 | 119806 | 6.46 | 666 | P < 0.001 |
| UDP-alpha-D-glucose:NAD+ 6-oxidoreductase | 1.1.1.22 | 341877 | 267968 | 6.45 | 666 | P < 0.001 |
| 4-O-beta-D-mannopyranosyl-D-glucopyranose:phosphate alpha-D-mannosyltransferase | 2.4.1.281 | 294563 | 221546 | 6.44 | 666 | P < 0.001 |
| O-acetyl-ADP-D-ribose carboxylesterase | 3.1.1.106 | 232755 | 178380 | 6.39 | 666 | P < 0.001 |
| 2′-deoxyribonucleoside-5′-diphosphate:thioredoxin-disulfide 2′-oxidoreductase | 1.17.4.1 | 355018 | 281224 | 6.39 | 666 | P < 0.001 |
| 3′-ribonucleotide phosphohydrolase | 3.1.3.6 | 303202 | 229568 | 6.38 | 666 | P < 0.001 |
| acetyl-CoA:alkane-alpha,omega-diamine N-acetyltransferase | 2.3.1.57 | 341806 | 269466 | 6.32 | 666 | P < 0.001 |
| 5,10-methylenetetrahydrofolate:glycine hydroxymethyltransferase | 2.1.2.1 | 356606 | 283241 | 6.30 | 666 | P < 0.001 |
| carbonic acid hydro-lyase (carbon-dioxide-forming) | 4.2.1.1 | 321295 | 248957 | 6.29 | 666 | P < 0.001 |
| L-isoleucine:tRNAIle ligase (AMP-forming) | 6.1.1.5 | 364094 | 292209 | 6.24 | 666 | P < 0.001 |
| DNA-6-O-methylguanine/DNA-4-O-methylthymine:[protein]-L-cysteine S-methyltransferase | 2.1.1.63 | 365615 | 293975 | 6.16 | 666 | P < 0.001 |
| NADH:hydroperoxide oxidoreductase | 1.11.1.26 | 323275 | 252045 | 6.11 | 666 | P < 0.001 |
| L-glutamate:tRNAGlu ligase (AMP-forming) | 6.1.1.17 | 321496 | 262066 | 6.08 | 666 | P < 0.001 |
| bleomycin hydrolase | 3.4.22.40 | 314461 | 243175 | 6.08 | 666 | P < 0.001 |
| L-cysteine-S-conjugate thiol-lyase (deaminating; 2-aminoprop-2-enoate-forming) | 4.4.1.13 | 363963 | 293237 | 6.08 | 666 | P < 0.001 |
| UDP-alpha-D-glucose:alpha-D-galactose-1-phosphate uridylyltransferase | 2.7.7.12 | 170072 | 129650 | 6.05 | 666 | P < 0.001 |
| peptidoglycan amidohydrolase | 3.5.1.28 | 363233 | 293964 | 5.99 | 666 | P < 0.001 |
| adenine-DNA deoxyribohydrolase (adenine-releasing) | 3.2.2.31 | 355124 | 285300 | 5.95 | 666 | P < 0.001 |
| D-phosphoglycerate 2,3-phosphomutase (2,3-diphosphoglycerate-dependent) | 5.4.2.11 | 319582 | 250208 | 5.95 | 666 | P < 0.001 |
| tRNA N6-(3-methylbut-2-en-1-yl)-adenine37:sulfur-(sulfur carrier),S-adenosyl-L-methionine C2-(methylsulfanyl)transferase | 2.8.4.3 | 370115 | 301224 | 5.93 | 666 | P < 0.001 |
| 10-formyltetrahydrofolate:5′-phosphoribosylglycinamide N-formyltransferase | 2.1.2.2 | 363374 | 295358 | 5.89 | 666 | P < 0.001 |
| peptidase Do | 3.4.21.107 | 276660 | 224637 | 5.87 | 666 | P < 0.001 |
| poly(deoxyribonucleotide)-3′-hydroxyl:5′-phospho-poly(deoxyribonucleotide) ligase (NAD+) | 6.5.1.2 | 372463 | 304412 | 5.85 | 666 | P < 0.001 |
| methionyl aminopeptidase | 3.4.11.18 | 372462 | 304426 | 5.85 | 666 | P < 0.001 |
| prenyl-diphosphate:adenine37 in tRNA prenyltransferase | 2.5.1.75 | 371863 | 303924 | 5.84 | 666 | P < 0.001 |
| ATP:pyridoxal 5′-phosphotransferase | 2.7.1.35 | 329032 | 261786 | 5.80 | 666 | P < 0.001 |
| L-rhamnose aldose-ketose-isomerase | 5.3.1.14 | 307393 | 241401 | 5.80 | 666 | P < 0.001 |
| peptide-methionine:thioredoxin-disulfide S-oxidoreductase [methionine (R)-S-oxide-forming] | 1.8.4.12 | 257798 | 203935 | 5.76 | 666 | P < 0.001 |
| deoxyribonuclease IV | 3.1.21.2 | 353617 | 287404 | 5.73 | 666 | P < 0.001 |
| alpha-L-rhamnoside rhamnohydrolase | 3.2.1.40 | 318053 | 252744 | 5.68 | 666 | P < 0.001 |
| ATP phosphohydrolase (ABC-type, Zn2+-importing) | 7.2.2.20 | 331254 | 264520 | 5.66 | 666 | P < 0.001 |
| L-rhamnulose-1-phosphate (S)-lactaldehyde-lyase (glycerone-phosphate-forming) | 4.1.2.19 | 307632 | 243590 | 5.63 | 666 | P < 0.001 |
| 6-deoxy-6-sulfo-D-fructose:D-glyceraldehyde-3-phosphate glyceronetransferase | 2.2.1.14 | 173 | 33 | 5.60 | 41 | P < 0.001 |
| ATP:L-rhamnulose 1-phosphotransferase | 2.7.1.5 | 319257 | 255641 | 5.50 | 666 | P < 0.001 |
| ATP:glycerol 3-phosphotransferase | 2.7.1.30 | 271460 | 223970 | 5.50 | 666 | P < 0.001 |
| (R)-S-lactoylglutathione methylglyoxal-lyase (isomerizing; glutathione-forming) | 4.4.1.5 | 341379 | 280079 | 5.36 | 666 | P < 0.001 |
| exodeoxyribonuclease V | 3.1.11.5 | 224710 | 181406 | 5.36 | 666 | P < 0.001 |
| NAD(H) phosphohydrolase | 3.6.1.22 | 335898 | 275329 | 5.26 | 666 | P < 0.001 |
| L-alanyl-D-glutamate epimerase | 5.1.1.20 | 211984 | 168065 | 5.24 | 666 | P < 0.001 |
| CTP:2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 2.7.7.60 | 342240 | 289619 | 5.12 | 666 | P < 0.001 |
| beta-D-fructofuranoside fructohydrolase | 3.2.1.26 | 204815 | 169106 | 4.91 | 666 | P < 0.001 |
| beta-lactam hydrolase | 3.5.2.6 | 328892 | 276729 | 4.76 | 666 | P < 0.001 |
| GTP 7,8-8,9-dihydrolase (diphosphate-forming) | 3.5.4.25 | 363191 | 310173 | 4.54 | 666 | P < 0.001 |
| D-ribulose 5-phosphate formate-lyase (L-3,4-dihydroxybutan-2-one 4-phosphate-forming) | 4.1.99.12 | 363190 | 310173 | 4.54 | 666 | P < 0.001 |
| 5-amino-6-(D-ribitylamino)uracil butanedionetransferase | 2.5.1.78 | 363021 | 310046 | 4.54 | 666 | P < 0.001 |
| 5-amino-6-(5-phospho-D-ribitylamino)uracil:NADP+ 1′-oxidoreductase | 1.1.1.193 | 356070 | 302730 | 4.53 | 666 | P < 0.001 |
| 2,5-diamino-6-hydroxy-4-(5-phospho-D-ribosylamino)pyrimidine 2-aminohydrolase | 3.5.4.26 | 356070 | 302730 | 4.53 | 666 | P < 0.001 |
| D-mannose-6-phosphate aldose-ketose-isomerase | 5.3.1.8 | 349959 | 298038 | 4.51 | 666 | P < 0.001 |
| 5′-ribonucleotide phosphohydrolase | 3.1.3.5 | 363417 | 312818 | 4.40 | 666 | P < 0.001 |
| peptidoglycan-N-acetylglucosamine amidohydrolase | 3.5.1.104 | 334065 | 289136 | 4.35 | 666 | P < 0.001 |
| fragilysin | 3.4.24.74 | 7726 | 2137 | 4.30 | 487 | P < 0.001 |
| UTP:alpha-D-glucose-1-phosphate uridylyltransferase | 2.7.7.9 | 180042 | 146829 | 4.27 | 666 | P < 0.001 |
| adenosylcobinamide-GDP:alpha-ribazole ribazoletransferase | 2.7.8.26 | 336101 | 289980 | 4.27 | 666 | P < 0.001 |
| Fe(II):oxygen oxidoreductase ([FeO(OH)]core-producing) | 1.16.3.2 | 343886 | 295642 | 4.23 | 666 | P < 0.001 |
| D-stereospecific aminopeptidase | 3.4.11.19 | 146732 | 114608 | 4.23 | 666 | P < 0.001 |
| D-glycerate:NAD+ oxidoreductase | 1.1.1.29 | 304790 | 258395 | 4.21 | 666 | P < 0.001 |
| ATP:dTMP phosphotransferase | 2.7.4.9 | 182727 | 149804 | 4.20 | 666 | P < 0.001 |
| ATP:UDP-N-acetyl-alpha-D-glucosamine 3′-phosphotransferase | 2.7.1.176 | 7304 | 3121 | 4.15 | 664 | P < 0.001 |
| L-histidinol:NAD+ oxidoreductase | 1.1.1.23 | 360776 | 312129 | 4.13 | 666 | P < 0.001 |
| queuosine34 in tRNA:acceptor oxidoreductase | 1.17.99.6 | 365803 | 318709 | 4.13 | 666 | P < 0.001 |
| L-lysine:tRNALys ligase (AMP-forming) | 6.1.1.6 | 372837 | 325856 | 4.10 | 666 | P < 0.001 |
| (R)-lactate hydro-lyase (adding N-acetyl-D-glucosamine 6-phosphate; N-acetylmuramate 6-phosphate-forming) | 4.2.1.126 | 315409 | 268045 | 4.06 | 666 | P < 0.001 |
| CoA-[4′-phosphopantetheine]:apo-[acyl-carrier protein] 4′-pantetheinephosphotransferase | 2.7.8.7 | 170465 | 139721 | 4.06 | 666 | P < 0.001 |
| D-altronate hydro-lyase (2-dehydro-3-deoxy-D-gluconate-forming) | 4.2.1.7 | 307568 | 262854 | 4.05 | 666 | P < 0.001 |
| (6R)-6beta-hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide 6-epimerase | 5.1.99.6 | 373721 | 327886 | 3.98 | 666 | P < 0.001 |
| UTP:L-glutamine amido-ligase (ADP-forming) | 6.3.4.2 | 369633 | 324526 | 3.96 | 666 | P < 0.001 |
| beta-D-4-deoxy-Delta4-GlcAp-(1->3)-beta-D-GalNAc6S hydrolase | 3.2.1.180 | 300245 | 258391 | 3.94 | 666 | P < 0.001 |
| D-erythro-1-(imidazol-4-yl)glycerol-3-phosphate hydro-lyase [3-(imidazol-4-yl)-2-oxopropyl-phosphate-forming] | 4.2.1.19 | 361792 | 316233 | 3.94 | 666 | P < 0.001 |
| 1-(5-phospho-beta-D-ribosyl)-ATP:diphosphate phospho-alpha-D-ribosyl-transferase | 2.4.2.17 | 361822 | 316271 | 3.94 | 666 | P < 0.001 |
| 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol CMP-lyase (cyclizing; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate-forming) | 4.6.1.12 | 375199 | 329906 | 3.93 | 666 | P < 0.001 |
| 1-(5-phospho-beta-D-ribosyl)-5-[(5-phospho-beta-D-ribosylamino)methylideneamino]imidazole-4-carboxamide aldose-ketose-isomerase | 5.3.1.16 | 361727 | 316261 | 3.93 | 666 | P < 0.001 |
| [poly-N-acetyl-D-glucosaminyl-(1->4)-(N-acetyl-D-muramoylpentapeptide)]-diphosphoundecaprenol:[N-acetyl-D-glucosaminyl-(1->4)-N-acetyl-D-muramoylpentapeptide]-diphosphoundecaprenol disaccharidetransferase | 2.4.1.129 | 367331 | 322381 | 3.93 | 666 | P < 0.001 |
| 5-[(5-phospho-1-deoxy-D-ribulos-1-ylamino)methylideneamino]-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamide D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate-lyase (L-glutamine-hydrolysing; 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamide-forming) | 4.3.2.10 | 363190 | 317925 | 3.91 | 666 | P < 0.001 |
| dTDP-alpha-D-glucose 4,6-hydro-lyase (dTDP-4-dehydro-6-deoxy-alpha-D-glucose-forming) | 4.2.1.46 | 363385 | 318389 | 3.90 | 666 | P < 0.001 |
| ATP:(d)CMP phosphotransferase | 2.7.4.25 | 370117 | 325801 | 3.88 | 666 | P < 0.001 |
| L-histidinol-phosphate:2-oxoglutarate aminotransferase | 2.6.1.9 | 372117 | 327934 | 3.85 | 666 | P < 0.001 |
| 5,10-methylenetetrahydrofolate,FADH2:dUMP C-methyltransferase | 2.1.1.148 | 143685 | 115112 | 3.81 | 666 | P < 0.001 |
| orotidine-5′-phosphate carboxy-lyase (UMP-forming) | 4.1.1.23 | 376841 | 333051 | 3.81 | 666 | P < 0.001 |
| N2-formyl-N1-(5-phospho-D-ribosyl)glycinamide:L-glutamine amido-ligase (ADP-forming) | 6.3.5.3 | 376690 | 332992 | 3.80 | 666 | P < 0.001 |
| alpha-D-galactoside galactohydrolase | 3.2.1.22 | 351869 | 309262 | 3.80 | 666 | P < 0.001 |
| oligonucleotidase | 3.1.13.3 | 361085 | 318401 | 3.79 | 666 | P < 0.001 |
| ATP:pseudouridine 5′-phosphotransferase | 2.7.1.83 | 143192 | 114831 | 3.78 | 666 | P < 0.001 |
| citrate(isocitrate) hydro-lyase (cis-aconitate-forming) | 4.2.1.3 | 370344 | 327004 | 3.78 | 666 | P < 0.001 |
| adenosine-3′(2′),5′-bisphosphate 3′(2′)-phosphohydrolase | 3.1.3.7 | 363418 | 321345 | 3.74 | 666 | P < 0.001 |
| adenylyl-molybdopterin:molybdate molybdate transferase (AMP-forming) | 2.10.1.1 | 155862 | 128309 | 3.71 | 666 | P < 0.001 |
| sn-glycerol-3-phosphate:NAD(P)+ 2-oxidoreductase | 1.1.1.94 | 370438 | 328604 | 3.70 | 666 | P < 0.001 |
| isocitrate:NADP+ oxidoreductase (decarboxylating) | 1.1.1.42 | 361230 | 319382 | 3.70 | 666 | P < 0.001 |
| short-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase | 1.3.8.1 | 140972 | 113231 | 3.70 | 666 | P < 0.001 |
| acetyl-CoA:oxaloacetate C-acetyltransferase [thioester-hydrolysing, (pro-S)-carboxymethyl-forming] | 2.3.3.1 | 362515 | 321195 | 3.65 | 666 | P < 0.001 |
| UDP-2,4-diacetamido-2,4,6-trideoxy-beta-L-altropyranose hydrolase | 3.6.1.57 | 5601 | 2056 | 3.64 | 644 | P < 0.001 |
| D-mannonate hydro-lyase (2-dehydro-3-deoxy-D-gluconate-forming) | 4.2.1.8 | 329439 | 288135 | 3.63 | 666 | P < 0.001 |
| ribonuclease M5 | 3.1.26.8 | 146813 | 119961 | 3.60 | 666 | P < 0.001 |
| (S)-4-hydroxymandelate:oxygen 1-oxidoreductase | 1.1.3.46 | 148308 | 122183 | 3.58 | 666 | P < 0.001 |
| S-adenosyl-L-methionine:tRNA (carboxymethyluridine34-5-O)-methyltransferase | 2.1.1.229 | 86 | 34 | 3.50 | 87 | P < 0.001 |
| 5,10-methylenetetrahydrofolate:tRNA (uracil54-C5)-methyltransferase | 2.1.1.74 | 146047 | 120006 | 3.48 | 666 | P < 0.001 |
| aldehyde:ferredoxin oxidoreductase | 1.2.7.5 | 140099 | 114474 | 3.47 | 666 | P < 0.001 |
| cobalt-precorrin-6B:NAD+ oxidoreductase | 1.3.1.106 | 150101 | 125120 | 3.41 | 666 | P < 0.001 |
| precorrin-6B:NADP+ oxidoreductase | 1.3.1.54 | 150101 | 125120 | 3.41 | 666 | P < 0.001 |
| (3S)-3-hydroxyacyl-CoA hydro-lyase | 4.2.1.17 | 146259 | 121343 | 3.40 | 666 | P < 0.001 |
| coproporphyrinogen-III:S-adenosyl-L-methionine oxidoreductase (decarboxylating) | 1.3.98.3 | 183567 | 156750 | 3.31 | 666 | P < 0.001 |
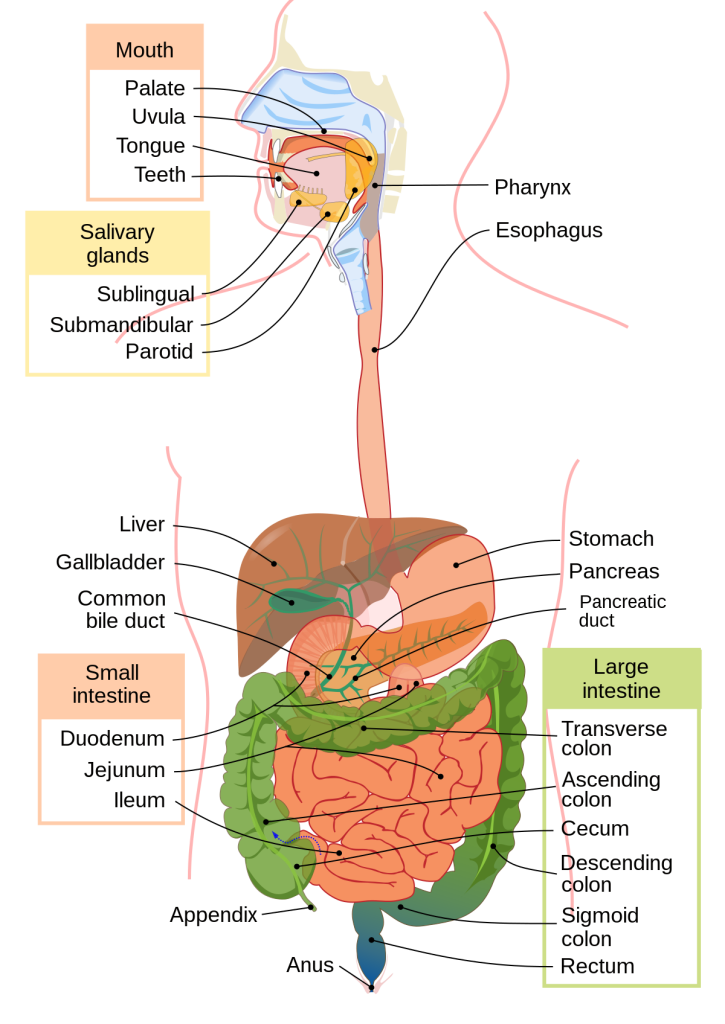



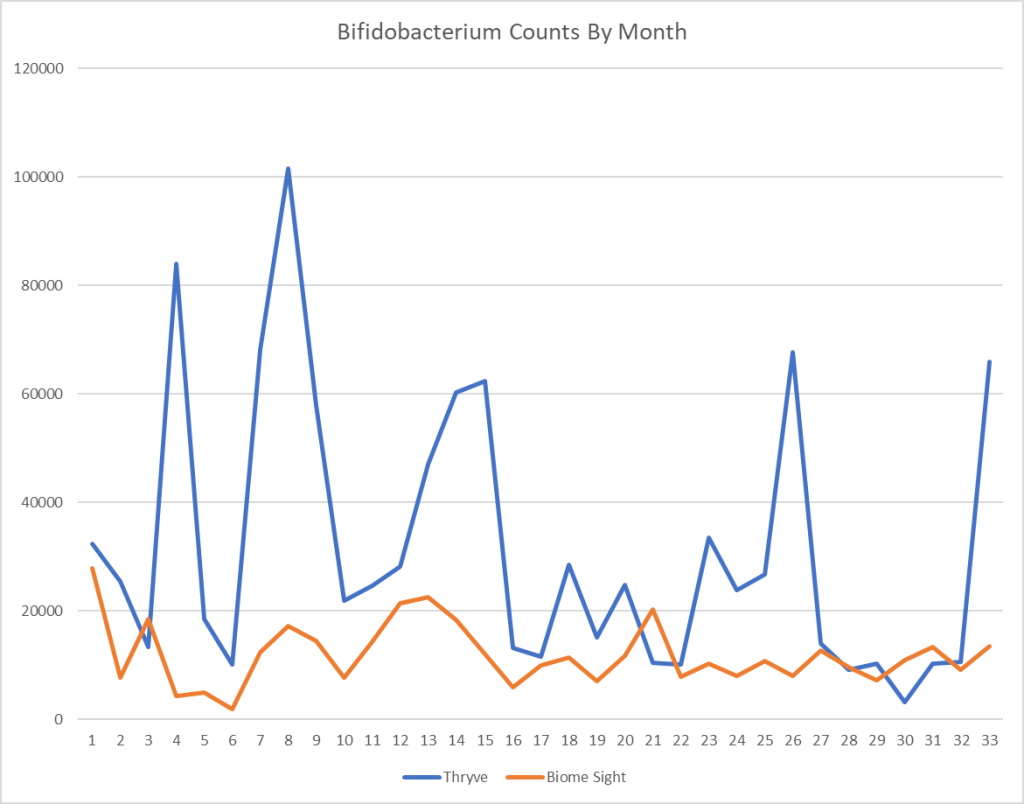






 Reversion of Gut Microbiota during the Recovery Phase in Patients with Asymptomatic or Mild COVID-19: Longitudinal Study.
Reversion of Gut Microbiota during the Recovery Phase in Patients with Asymptomatic or Mild COVID-19: Longitudinal Study.




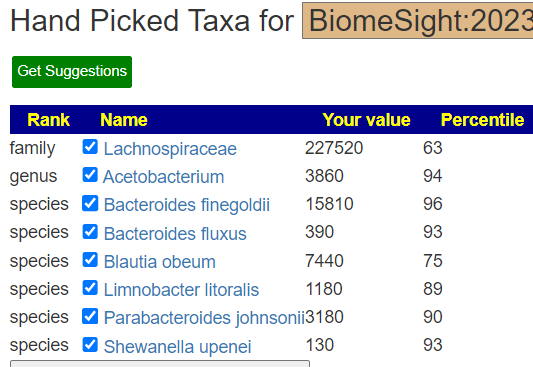
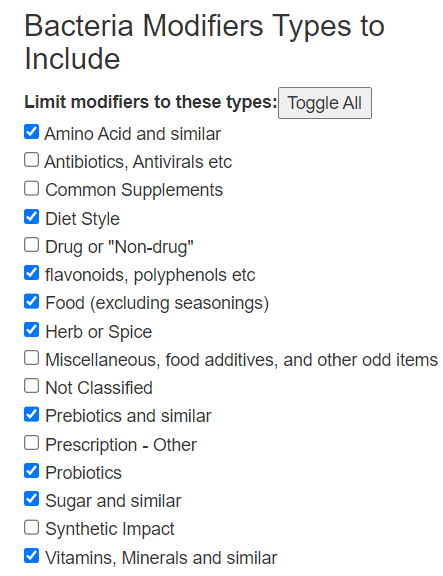
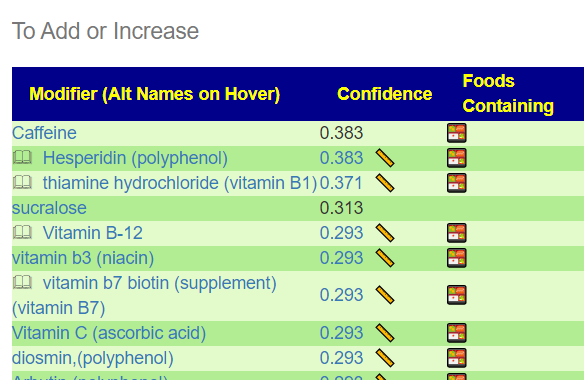


Recent Comments