This is a common symptom for many people with Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). This is reported often in samples, and thus being examined if it reaches our threshold for inclusion as defined in A new specialized selection of suggestions links. It does. We are not being specific about the type of allergy.
Sub-Series Study Populations:
We have 5 symptom annotations that this sub-series are examining, I tried different combination to see which resulted in a higher z-score to identify probable siblings (in a statistical sense)
- New food sensitivities – 12.1 z-score
- Medication sensitivities -8.8 z-score
- Combined, dropped to 6.4
- Alcohol intolerance – 8.6 z-score
- with Medication sensitivities 8.6 z-score and less bacteria
- Allergic Rhinitis (Hay Fever) – 8.6 z-score
- When combined with any of the above, a major drop of z-scores
- Allergies – 9.2 z-score
- With new food sensitivities – 9.9 z-score
- When combined with any of the other, a major drop of z-scores
We have broken this down into 3 sub-groups of microbiome shifts:
- Allergies and Food Sensitivity
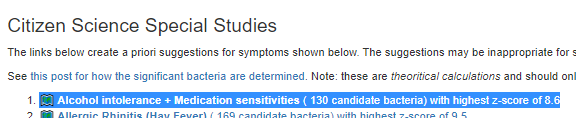
- Alcohol intolerance + Medication sensitivities (this study).
- Allergic Rhinitis (Hay Fever)
| Symptom | Reference | Study |
| Alcohol intolerance + Medication sensitivities | 1144 | 59 |
- Bacteria Detected with z-score > 2.6: found 130 items, highest value was 8.6
- Enzymes Detected with z-score > 2.6: found 507 items, highest value was 7.9
- Compound Detected with z-score > 2.6: found No items
The bacteria that was most significant was NOT Prevotella copri (species) which we saw with Special Studies: Allergies And Food Sensitivity.
Interesting Significant Bacteria
All bacteria found significant (except 1) had too low levels. This is a common pattern found with these studies, it is not “bad bacteria bogie man bacteria” but an absence of “upstanding citizens bacteria”. The signature is very interesting because it is specific Bifidobacterium species — despite this, the Bifidobacterium genus was not found to be statistically significant. This drops us into the odd space of general Bifidobacterium probiotics likely not be effective, but specific strains may be(unfortunately those strains are not available as probiotics). There is one retail probiotics that contains Enterbacter, General Biotics/Equilibrium which give an experiment to try.
| Bacteria | Reference Mean | Study | Z-Score |
| Bifidobacterium gallicum (species) | 3698 | 136 | 8.6 |
| Bifidobacterium catenulatum subsp. kashiwanohense (subspecies) | 316 | 55 | 6.9 |
| Bifidobacterium kashiwanohense PV20-2 (strain) | 315 | 55 | 6.8 |
| Enterobacter (genus) | 1361 | 127 | 5.6 |
| Bifidobacterium angulatum (species) | 183 | 20 | 5.2 |
| Gammaproteobacteria (class) | 14010 | 5636 | 5 |
| Aeromonadaceae (family) | 157 | 25 | 5 |
Interesting Enzymes
All 507 enzymes found significant had too low levels. This places it into the same class of massive enzyme disruptions as seen in Special Studies: Depression . As with that one, apologies for the massive list — I am keeping to my practice for these studies of listing everything with 5.0 z-score or higher.
| Enzyme | Reference Mean | Study Mean | Z-Score |
| 2-acetylphloroglucinol C-acetyltransferase (2.3.1.272) | 177 | 28 | 7.9 |
| [cysteine desulfurase]-S-sulfanyl-L-cysteine:[molybdopterin-synthase sulfur-carrier protein]-Gly-Gly sulfurtransferase (2.8.1.11) | 5231 | 1805 | 7.7 |
| S-adenosyl-L-methionine:tellurite methyltransferase (2.1.1.265) | 5238 | 1938 | 7 |
| decenoyl-[acyl-carrier protein] Delta2-trans-Delta3-cis-isomerase (5.3.3.14) | 6010 | 2276 | 6.9 |
| tRNA-uridine65 uracil mutase (5.4.99.26) | 4052 | 1226 | 6.8 |
| coproporphyrinogen:oxygen oxidoreductase (decarboxylating) (1.3.3.3) | 3163 | 746 | 6.8 |
| donor:hydrogen-peroxide oxidoreductase (1.11.1.21) | 3030 | 672 | 6.7 |
| 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase (5.3.3.18) | 3097 | 1059 | 6.7 |
| 3-hydroxybutanoyl-CoA 3-epimerase (5.1.2.3) | 2951 | 696 | 6.7 |
| ATP:nicotinamide-nucleotide adenylyltransferase (2.7.7.1) | 5076 | 2200 | 6.7 |
| ATP:1-(beta-D-ribofuranosyl)-nicotinamide 5′-phosphotransferase (2.7.1.22) | 5025 | 2187 | 6.6 |
| isocitrate glyoxylate-lyase (succinate-forming) (4.1.3.1) | 2955 | 730 | 6.6 |
| D-amino acid:quinone oxidoreductase (deaminating) (1.4.5.1) | 2485 | 686 | 6.5 |
| L-methionine:2-oxo-acid aminotransferase (2.6.1.88) | 2759 | 652 | 6.5 |
| 2-(glutathione-S-yl)-hydroquinone:glutathione oxidoreductase (1.8.5.7) | 3140 | 748 | 6.5 |
| D-glucose:ubiquinone oxidoreductase (1.1.5.2) | 2200 | 462 | 6.5 |
| methanesulfonate,FMNH2:oxygen oxidoreductase (1.14.14.34) | 2239 | 489 | 6.5 |
| alkanesulfonate,FMNH2:oxygen oxidoreductase (1.14.14.5) | 2239 | 489 | 6.5 |
| glutathione:NADP+ oxidoreductase (1.8.1.7) | 5947 | 2494 | 6.5 |
| triphosphate phosphohydrolase (3.6.1.25) | 4494 | 2012 | 6.4 |
| S-(hydroxymethyl)glutathione:NAD+ oxidoreductase (1.1.1.284) | 2862 | 759 | 6.4 |
| (S)-3-hydroxyacyl-CoA:NAD+ oxidoreductase (1.1.1.35) | 3374 | 926 | 6.4 |
| ferredoxin:NAD+ oxidoreductase (1.18.1.3) | 1992 | 455 | 6.4 |
| acyl-CoA:sn-glycerol-3-phosphate 1-O-acyltransferase (2.3.1.15) | 3270 | 1139 | 6.4 |
| S-adenosyl-L-methionine:tRNA (cytidine32/uridine32-2′-O)-methyltransferase (2.1.1.200) | 3316 | 1093 | 6.3 |
| [50S ribosomal protein L16]-L-Arg81,2-oxoglutarate:oxygen oxidoreductase (3R-hydroxylating) (1.14.11.47) | 3334 | 1114 | 6.3 |
| choline:acceptor 1-oxidoreductase (1.1.99.1) | 2748 | 562 | 6.3 |
| acetyl-CoA:glyoxylate C-acetyltransferase [(S)-malate-forming] (2.3.3.9) | 3116 | 950 | 6.3 |
| siroheme ferro-lyase (sirohydrochlorin-forming) (4.99.1.4) | 2565 | 539 | 6.2 |
| deoxyribocyclobutadipyrimidine pyrimidine-lyase (4.1.99.3) | 4156 | 1337 | 6.2 |
| acetyl-CoA:[elongator tRNAMet]-cytidine34 N4-acetyltransferase (ATP-hydrolysing) (2.3.1.193) | 3309 | 1094 | 6.2 |
| 7,8-dihydroneopterin 3′-triphosphate diphosphohydrolase (3.6.1.67) | 3607 | 1299 | 6.2 |
| n/a (3.4.11.23) | 3312 | 1083 | 6.2 |
| acetyl-CoA:dTDP-4-amino-4,6-dideoxy-alpha-D-galactose N-acetyltransferase (2.3.1.210) | 2766 | 673 | 6.2 |
| ATP:N-acetyl-D-glucosamine 6-phosphotransferase (2.7.1.59) | 3263 | 1071 | 6.2 |
| L-2,4-diaminobutanoate carboxy-lyase (propane-1,3-diamine-forming) (4.1.1.86) | 2127 | 304 | 6.2 |
| chorismate pyruvate-lyase (4-hydroxybenzoate-forming) (4.1.3.40) | 2596 | 567 | 6.2 |
| protein dithiol:quinone oxidoreductase (disulfide-forming) (1.8.5.9) | 3557 | 1276 | 6.2 |
| fatty acyl-[acyl-carrier protein]:alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-(acyl)-[lipid IVA] O-acyltransferase (2.3.1.243) | 3322 | 1097 | 6.2 |
| ATP:L-threonine O3-phosphotransferase (2.7.1.177) | 2358 | 350 | 6.1 |
| ditrans-octacis-undecaprenyl-diphosphate:alpha-D-Kdo-(2->4)-alpha-D-Kdo-(2->6)-lipid-A phosphotransferase (2.7.4.29) | 2852 | 1008 | 6.1 |
| D-sorbitol-6-phosphate:NAD+ 2-oxidoreductase (1.1.1.140) | 2447 | 677 | 6.1 |
| diacylglycerol-3-phosphate phosphohydrolase (3.1.3.4) | 3284 | 1105 | 6.1 |
| 1,2-diacyl-sn-glycerol 3-phosphate phosphohydrolase (3.1.3.81) | 3284 | 1105 | 6.1 |
| (R)-lactate:quinone 2-oxidoreductase (1.1.5.12) | 2328 | 510 | 6.1 |
| (S)-lactate:ferricytochrome-c 2-oxidoreductase (1.1.2.3) | 2368 | 544 | 6.1 |
| methyl DNA-base, 2-oxoglutarate:oxygen oxidoreductase (formaldehyde-forming) (1.14.11.33) | 2304 | 475 | 6.1 |
| ATP:D-ribulose-5-phosphate 1-phosphotransferase (2.7.1.19) | 2435 | 543 | 6.1 |
| (2S,3S)-2,3-dihydro-2,3-dihydroxybenzoate:NAD+ oxidoreductase (1.3.1.28) | 2240 | 476 | 6.1 |
| D-mannitol-1-phosphate:NAD+ 5-oxidoreductase (1.1.1.17) | 2672 | 727 | 6 |
| 2-O-(alpha-D-mannopyranosyl)-D-glycerate D-mannohydrolase (3.2.1.170) | 1248 | 396 | 6 |
| S-formylglutathione hydrolase (3.1.2.12) | 2402 | 554 | 6 |
| gamma-L-glutamyl-L-cysteinyl-glycine:spermidine amidase (3.5.1.78) | 3189 | 1059 | 6 |
| gamma-L-glutamyl-L-cysteinyl-glycine:spermidine ligase (ADP-forming) [spermidine is numbered so that atom N-1 is in the amino group of the aminopropyl part of the molecule] (6.3.1.8) | 3189 | 1059 | 6 |
| S-adenosyl-L-methionine:23S rRNA (uracil747-C5)-methyltransferase (2.1.1.189) | 3257 | 1112 | 6 |
| S-adenosyl-L-methionine:uridine in tRNA 3-[(3S)-3-amino-3-carboxypropyl]transferase (2.5.1.25) | 1704 | 303 | 6 |
| succinate-semialdehyde:NAD+ oxidoreductase (1.2.1.24) | 2416 | 449 | 6 |
| Fe(II)-siderophore:NADP+ oxidoreductase (1.16.1.9) | 2344 | 447 | 6 |
| ATP phosphohydrolase (ABC-type, taurine-importing) (7.6.2.7) | 2185 | 521 | 6 |
| geraniol:NADP+ oxidoreductase (1.1.1.183) | 2470 | 466 | 6 |
| ATP:N-acyl-D-mannosamine 6-phosphotransferase (2.7.1.60) | 3204 | 1098 | 5.9 |
| L-serine:[L-seryl-carrier protein] ligase (AMP-forming) (6.2.1.72) | 2199 | 454 | 5.9 |
| galactarate hydro-lyase (5-dehydro-4-deoxy-D-glucarate-forming) (4.2.1.42) | 3015 | 963 | 5.9 |
| propanoyl-CoA:phosphate propanoyltransferase (2.3.1.222) | 1738 | 358 | 5.9 |
| ATP:thiamine phosphotransferase (2.7.1.89) | 2372 | 492 | 5.9 |
| ATP phosphohydrolase (ABC-type, putrescine-importing) (7.6.2.16) | 2302 | 503 | 5.9 |
| ubiquinol:oxygen oxidoreductase (superoxide-forming) (1.10.3.17) | 2232 | 475 | 5.9 |
| S-adenosyl-L-methionine:23S rRNA (guanine1835-N2)-methyltransferase (2.1.1.174) | 2258 | 485 | 5.9 |
| riboflavin:NAD(P)+ oxidoreductase (1.5.1.41) | 2323 | 491 | 5.9 |
| D-erythrose 4-phosphate:NAD+ oxidoreductase (1.2.1.72) | 2267 | 485 | 5.9 |
| CDP-diacylglycerol phosphatidylhydrolase (3.6.1.26) | 2239 | 499 | 5.9 |
| D-glucarate hydro-lyase (5-dehydro-4-deoxy-D-glucarate-forming) (4.2.1.40) | 2953 | 1125 | 5.9 |
| 6-phospho-D-gluconate hydro-lyase (2-dehydro-3-deoxy-6-phospho-D-gluconate-forming) (4.2.1.12) | 2260 | 495 | 5.8 |
| ATP:propanoate phosphotransferase (2.7.2.15) | 2215 | 469 | 5.8 |
| isochorismate pyruvate-hydrolase (3.3.2.1) | 2164 | 473 | 5.8 |
| (3Z/3E)-alk-3-enoyl-CoA (2E)-isomerase (5.3.3.8) | 2225 | 487 | 5.8 |
| ATP phosphohydrolase (ABC-type, L-arabinose-importing) (7.5.2.12) | 2214 | 473 | 5.8 |
| n/a (3.4.24.55) | 2279 | 492 | 5.8 |
| diacylphosphatidylethanolamine:alpha-D-Kdo-(2->4)-alpha-D-Kdo-(2->6)-lipid-A 7”-phosphoethanolaminetransferase (2.7.8.42) | 2371 | 500 | 5.8 |
| 2,3-dihydroxybenzoate:L-serine ligase (6.3.2.14) | 2179 | 611 | 5.8 |
| 2-O-(alpha-D-glucopyranosyl)-D-glycerate:phosphate alpha-D-glucosyltransferase (configuration-retaining) (2.4.1.352) | 1217 | 249 | 5.8 |
| pyrimidine-5′-nucleotide phosphoribo(deoxyribo)hydrolase (3.2.2.10) | 2222 | 495 | 5.8 |
| 6-sulfo-alpha-D-quinovosyl diacylglycerol 6-sulfo-D-quinovohydrolase (3.2.1.199) | 1113 | 222 | 5.8 |
| (9Z)-hexadec-9-enoyl-[acyl-carrier protein]:Kdo2-lipid IVA O-palmitoleoyltransferase (2.3.1.242) | 2356 | 502 | 5.8 |
| [RNA]-adenosine-cytidine 5′-hydroxy-adenosoine ribonucleotide-3′-[RNA fragment]-lyase (cyclicizing; [RNA fragment]-3′-cytidine-2′,3′-cyclophosphate-forming and hydrolysing) (4.6.1.21) | 2312 | 470 | 5.8 |
| ATP phosphohydrolase (ABC-type, D-allose-importing) (7.5.2.8) | 2146 | 431 | 5.8 |
| 6-methoxy-3-methyl-2-(all-trans-polyprenyl)-1,4-benzoquinol,acceptor:oxygen oxidoreductase (5-hydroxylating) (1.14.99.60) | 2180 | 540 | 5.8 |
| NADPH:NAD+ oxidoreductase (Si-specific) (1.6.1.1) | 2204 | 490 | 5.8 |
| taurine, 2-oxoglutarate:oxygen oxidoreductase (sulfite-forming) (1.14.11.17) | 2122 | 496 | 5.8 |
| (deoxy)cytidine 5′-triphosphate diphosphohydrolase (3.6.1.65) | 2307 | 500 | 5.7 |
| 2-(all-trans-polyprenyl)phenol,NADPH:oxygen oxidoreductase (6-hydroxylating) (1.14.13.240) | 2178 | 535 | 5.7 |
| dTDP-N-acetyl-alpha-D-fucose:N-acetyl-beta-D-mannosaminouronyl-(1->4)-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol N-acetylfucosaminyltransferase (2.4.1.325) | 2308 | 507 | 5.7 |
| ATP:[isocitrate dehydrogenase (NADP+)] phosphotransferase (2.7.11.5) | 2142 | 516 | 5.7 |
| 2,3-dihydroxybenzoate:[aryl-carrier protein] ligase (AMP-forming) (6.2.1.71) | 2118 | 604 | 5.7 |
| 5,6,7,8-tetrahydromonapterin:NADP+ oxidoreductase (1.5.1.50) | 2159 | 476 | 5.7 |
| glutathione gamma-glutamyl cyclotransferase (5-oxo-L-proline producing) (4.3.2.7) | 2158 | 500 | 5.7 |
| iron(III)-enterobactin hydrolase (3.1.1.108) | 2204 | 491 | 5.7 |
| N,N’-diacetylchitobiose acetylhydrolase (3.5.1.105) | 2304 | 508 | 5.7 |
| N-succinyl-L-glutamate amidohydrolase (3.5.1.96) | 2172 | 518 | 5.7 |
| protein-Npi-phospho-L-histidine:N-acetyl-D-muramate Npi-phosphotransferase (2.7.1.192) | 2283 | 474 | 5.7 |
| 4-alpha-D-[(1->4)-alpha-D-glucano]trehalose glucanohydrolase (trehalose-producing) (3.2.1.141) | 1335 | 181 | 5.7 |
| acetyl-CoA:N6-hydroxy-L-lysine 6-acetyltransferase (2.3.1.102) | 708 | 159 | 5.6 |
| 4-aminobutanoate:2-oxoglutarate aminotransferase (2.6.1.19) | 3600 | 1155 | 5.6 |
| ATP:(S)-4,5-dihydroxypentane-2,3-dione 5-phosphotransferase (2.7.1.189) | 1415 | 220 | 5.6 |
| inosine/xanthosine 5′-triphosphate phosphohydrolase (3.6.1.73) | 2071 | 496 | 5.6 |
| 6-deoxy-6-sulfofructose-1-phosphate 2-hydroxy-3-oxopropane-1-sulfonate-lyase (glycerone-phosphate-forming) (4.1.2.57) | 1346 | 295 | 5.6 |
| protein-Npi-phospho-L-histidine:2-O-alpha-mannopyranosyl-D-glycerate Npi-phosphotransferase (2.7.1.195) | 1163 | 234 | 5.6 |
| 2,3-dihydroxypropane-1-sulfonate:NAD+ 3-oxidoreductase (1.1.1.373) | 1341 | 295 | 5.6 |
| 4-aminobutanal:NAD+ 1-oxidoreductase (1.2.1.19) | 2237 | 491 | 5.6 |
| N-succinyl-L-glutamate 5-semialdehyde:NAD+ oxidoreductase (1.2.1.71) | 2135 | 535 | 5.6 |
| protein-dithiol:NAD(P)+ oxidoreductase (1.8.1.8) | 2151 | 540 | 5.6 |
| succinyl-CoA:acetyl-CoA C-succinyltransferase (2.3.1.174) | 1445 | 205 | 5.6 |
| succinyl-CoA:L-arginine N2-succinyltransferase (2.3.1.109) | 2135 | 536 | 5.6 |
| 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde:NADP+ oxidoreductase (1.2.1.91) | 1461 | 221 | 5.6 |
| 2-oxepin-2(3H)-ylideneacetyl-CoA hydrolase (3.3.2.12) | 1461 | 221 | 5.6 |
| N2-succinyl-L-arginine iminohydrolase (decarboxylating) (3.5.3.23) | 2134 | 536 | 5.6 |
| ATP phosphohydrolase (ABC-type, nitrate-importing) (7.3.2.4) | 1523 | 142 | 5.6 |
| 7,8-dihydroneopterin 3′-triphosphate 2′-epimerase (5.1.99.7) | 1928 | 468 | 5.5 |
| N4-acetylcytidine amidohydrolase (3.5.1.135) | 2966 | 1096 | 5.5 |
| N2-succinyl-L-ornithine:2-oxoglutarate 5-aminotransferase (2.6.1.81) | 2254 | 677 | 5.5 |
| (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase (5.4.99.15) | 1319 | 183 | 5.5 |
| ATP:2-dehydro-3-deoxy-D-galactonate 6-phosphotransferase (2.7.1.58) | 1536 | 299 | 5.5 |
| 2-dehydro-3-deoxy-6-phospho-D-galactonate D-glyceraldehyde-3-phospho-lyase (pyruvate-forming) (4.1.2.21) | 1538 | 299 | 5.5 |
| (S)-ureidoglycolate:NAD+ oxidoreductase (1.1.1.350) | 1729 | 436 | 5.5 |
| ATP:D-glyceraldehyde 3-phosphotransferase (2.7.1.28) | 1512 | 115 | 5.5 |
| ATP:glycerone phosphotransferase (2.7.1.29) | 1512 | 115 | 5.5 |
| FAD AMP-lyase (riboflavin-cyclic-4′,5′-phosphate-forming) (4.6.1.15) | 1512 | 115 | 5.5 |
| tRNA 2-(methylsulfanyl)-N6-prenyladenosine37,donor:oxygen 4-oxidoreductase (trans-hydroxylating) (1.14.99.69) | 1565 | 158 | 5.5 |
| S-methyl-5′-thioadenosine:phosphate S-methyl-5-thio-alpha-D-ribosyl-transferase (2.4.2.28) | 3673 | 1543 | 5.4 |
| primary-amine:oxygen oxidoreductase (deaminating) (1.4.3.21) | 1495 | 217 | 5.4 |
| UDP-4-amino-4-deoxy-alpha-L-arabinose:ditrans,octacis-undecaprenyl phosphate 4-amino-4-deoxy-alpha-L-arabinosyltransferase (2.4.2.53) | 2107 | 573 | 5.4 |
| (2S)-2-hydroxy-3,4-dioxopentyl phosphate aldose-ketose-isomerase (5.3.1.32) | 1529 | 259 | 5.4 |
| acyl-CoA:acetyl-CoA C-acyltransferase (2.3.1.16) | 3869 | 1876 | 5.4 |
| 2,3-didehydroadipoyl-CoA:acetyl-CoA C-didehydroadipoyltransferase (double bond migration) (2.3.1.223) | 1516 | 224 | 5.4 |
| 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase (2.1.2.13) | 2048 | 470 | 5.4 |
| UDP-alpha-D-glucuronate:NAD+ oxidoreductase (decarboxylating) (1.1.1.305) | 2048 | 470 | 5.4 |
| 4-amino-4-deoxy-alpha-L-arabinopyranosyl ditrans,octacis-undecaprenyl-phosphate:lipid IVA 4-amino-4-deoxy-L-arabinopyranosyltransferase (2.4.2.43) | 2044 | 473 | 5.4 |
| 5-(methylsulfanyl)-D-ribulose-1-phosphate 4-hydro-lyase [5-(methylsulfanyl)-2,3-dioxopentyl-phosphate-forming] (4.2.1.109) | 1552 | 181 | 5.3 |
| UDP-4-amino-4-deoxy-beta-L-arabinose:2-oxoglutarate aminotransferase (2.6.1.87) | 2070 | 480 | 5.3 |
| phenylacetyl-CoA:oxygen oxidoreductase (1,2-epoxidizing) (1.14.13.149) | 1375 | 278 | 5.3 |
| ATP phosphohydrolase (ABC-type, D-xylose-transporting) (7.5.2.10) | 1473 | 308 | 5.3 |
| 5-(methylsulfanyl)-2,3-dioxopentyl-phosphate phosphohydrolase (isomerizing) (3.1.3.77) | 1689 | 121 | 5.3 |
| N-benzoylamino-acid amidohydrolase (3.5.1.32) | 1712 | 408 | 5.3 |
| phenylacetaldehyde:NAD+ oxidoreductase (1.2.1.39) | 1391 | 271 | 5.3 |
| L-glutamate:putrescine ligase (ADP-forming) (6.3.1.11) | 1902 | 448 | 5.3 |
| iron(III)-salmochelin complex hydrolase (3.1.1.109) | 727 | 159 | 5.3 |
| 2-O-(alpha-D-mannosyl)-3-phosphoglycerate phosphohydrolase (3.1.3.70) | 2061 | 498 | 5.3 |
| 4-(gamma-L-glutamylamino)butanoate amidohydrolase (3.5.1.94) | 1914 | 445 | 5.3 |
| GDP-alpha-D-mannuronate:mannuronan D-mannuronatetransferase (2.4.1.33) | 154 | 39 | 5.2 |
| [mannuronan]-beta-D-mannuronate 5-epimerase (5.1.3.37) | 154 | 39 | 5.2 |
| 3-(indol-3-yl)pyruvate carboxy-lyase [(2-indol-3-yl)acetaldehyde-forming] (4.1.1.74) | 1802 | 138 | 5.2 |
| quinate:quinol 3-oxidoreductase (1.1.5.8) | 1523 | 124 | 5.2 |
| betaine-aldehyde:NAD+ oxidoreductase (1.2.1.8) | 2057 | 434 | 5.2 |
| catechol:oxygen 2,3-oxidoreductase (ring-opening) (1.13.11.2) | 3240 | 1416 | 5.2 |
| 1,2-dihydroxy-5-(methylsulfanyl)pent-1-en-3-one:oxygen oxidoreductase (formate- and CO-forming) (1.13.11.53) | 1714 | 283 | 5.2 |
| 1,2-dihydroxy-5-(methylsulfanyl)pent-1-en-3-one:oxygen oxidoreductase (formate-forming) (1.13.11.54) | 1714 | 283 | 5.2 |
| ATP phosphohydrolase (ABC-type, thiamine-importing) (7.6.2.15) | 3503 | 1300 | 5.2 |
| citrate:N6-acetyl-N6-hydroxy-L-lysine ligase (AMP-forming) (6.3.2.38) | 797 | 193 | 5.1 |
| RX:glutathione R-transferase (2.5.1.18) | 2968 | 1025 | 5.1 |
| N2-citryl-N6-acetyl-N6-hydroxy-L-lysine:N6-acetyl-N6-hydroxy-L-lysine ligase (AMP-forming) (6.3.2.39) | 795 | 193 | 5.1 |
| putrescine:2-oxoglutarate aminotransferase (2.6.1.82) | 3168 | 1490 | 5.1 |
| UDP-alpha-D-glucose:enterobactin 5′-C-beta-D-glucosyltransferase (configuration-inverting) (2.4.1.369) | 807 | 200 | 5.1 |
| 4-hydroxybutanoate:NAD+ oxidoreductase (1.1.1.61) | 2322 | 607 | 5.1 |
| D-xylonate hydro-lyase (2-dehydro-3-deoxy-D-arabinonate-forming) (4.2.1.82) | 621 | 178 | 5 |
| D-glycerate:NAD(P)+ oxidoreductase (1.1.1.60) | 1888 | 516 | 5 |
| galactitol-1-phosphate:NAD+ oxidoreductase (1.1.1.251) | 997 | 277 | 5 |
| alkylmercury mercury(II)-lyase (alkane-forming) (4.99.1.2) | 577 | 115 | 5 |
| (S)-2-hydroxyglutarate:quinone oxidoreductase (1.1.5.13) | 2192 | 670 | 5 |
| (R)-3-hydroxyacyl-CoA:NADP+ oxidoreductase (1.1.1.36) | 523 | 154 | 5 |
Bottom Line
These two symptoms tend to be ignored by the medical establishment. There are studies on the microbiome impacting the efficiency of prescription drugs but not sensitivities or hyper-reactivity. This means that this post is likely the first study on these in existence.
Social Thread
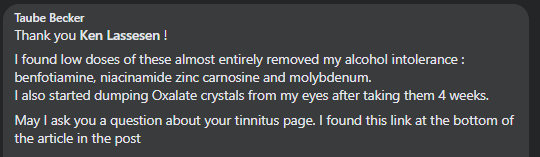
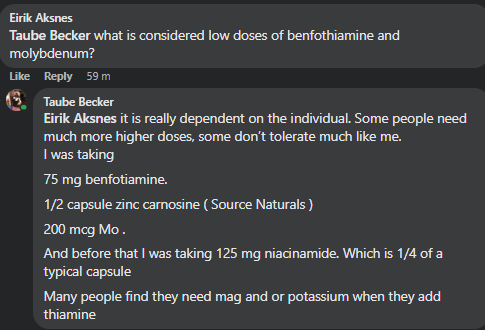
“There is one retail probiotics that contains Enterbacter, General Biotics/Equilibrium which give an experiment to try.”
As far as I know, General Biotics Equilibrium is not available anymore. I haven’t been able to place an order for some time on their website, they haven’t been active on any of their social media for more than a year, don’t respond to emails and in other stores it’s out of stock. Rather unfortunate