There are two approaches to identifying bacteria associated with a group of symptoms:
- UNION — you just join the bacteria associated with each symptom into a single list. This is often done when there is not sufficient data. It’s simple to do.
- INTERSECTION — this identifies all people with the same combination of symptoms and then identify what is associated. This requires statistical computations to be done each time.
Using the technic for statistical significance describe in my prior post, Symptom associated Bacteria, Compounds and Enzymes, I have successfully implemented it for samples contributed to my Citizen Science web site. The site is open data so you can replicate results.
The video below is a quick walkthrough. What is interesting to note is that the number of significant bacteria can increase as more symptoms are added. Why? because you are filtering out noise from the bacteria.
You can also have bacteria appearing that were not in the prior list by adding one more symptom. Example below.
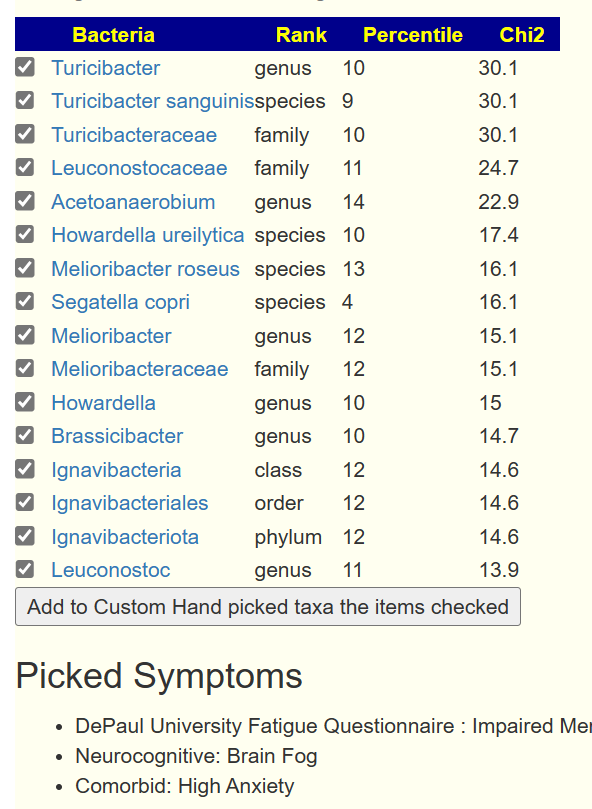
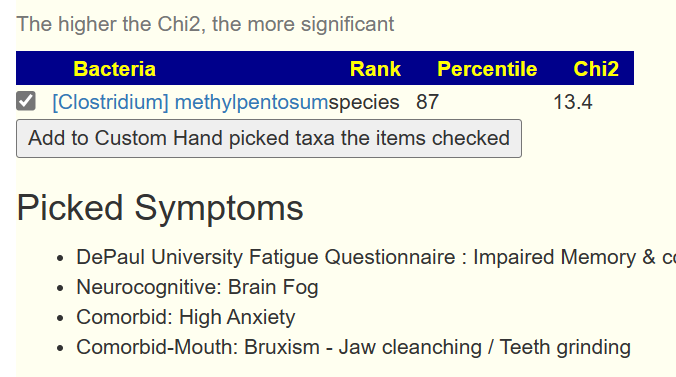
Bottom Line
With a large enough sample and enough characteristics recorded, you can drill down into a lot more data using the appropriate statistical techniques.
Recent Comments