Prior posts for this person are linked below
- The ME/CFS Quest for Health [May 2025]
- Ongoing ME/CFS Journey [Jan 2025]
- ME/CFS Continues Improvement + Lab Read Quality Issues [Feb 2024]
- Update on ME/CFS Person [Sep 2023]
- Follow up Microbiome Analysis from a prior post[Apr 2022]
- Rosacea, Circulation and mild CFS [Dec 2021]
I am going to review using the traditional analysis. My initial impression is that suggested retesting and plotting the next course correction was missed. As with sailing a boat, this can sail a person to an unintended spot. The last section is trying some work in progress on his sample(s). This is experimental work which I have high hope on yielding much finer identification of the critical bacteria that should be addressed.
Analysis
My usual starting point for multiple tests is compare forecasts of symptoms: New sample is 2025-11, old sample is 2025-03. Things have gotten worse.
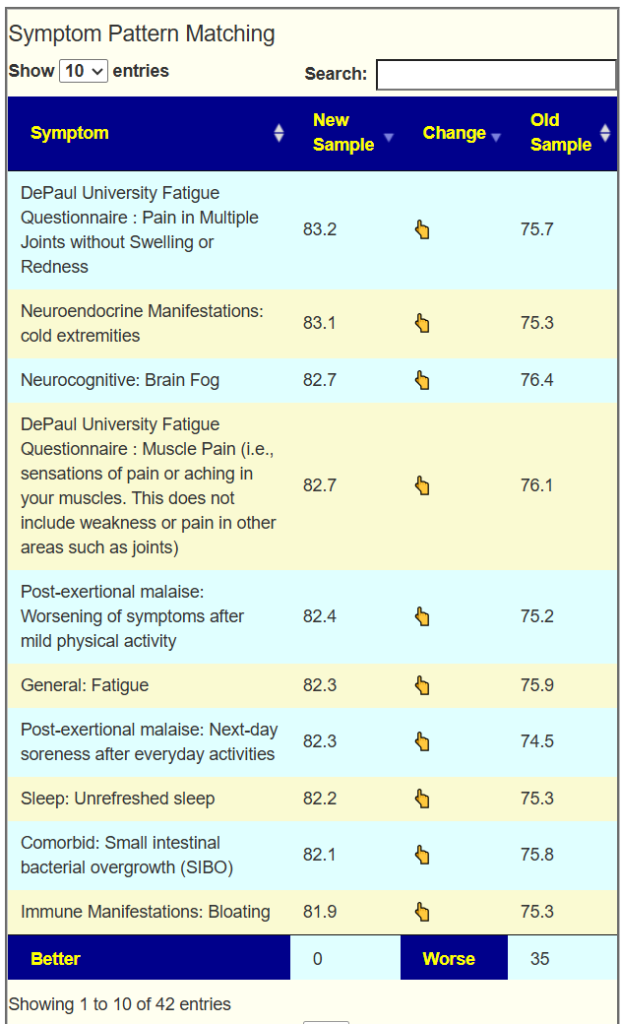
I decide to compare 2025-11 to 2024-12 and see the latest sample is still worse, but not as bad. In other words, the gains made over the summer has been lost. This person is in a northern climate so seasonal variation could (theoretically) be significant.
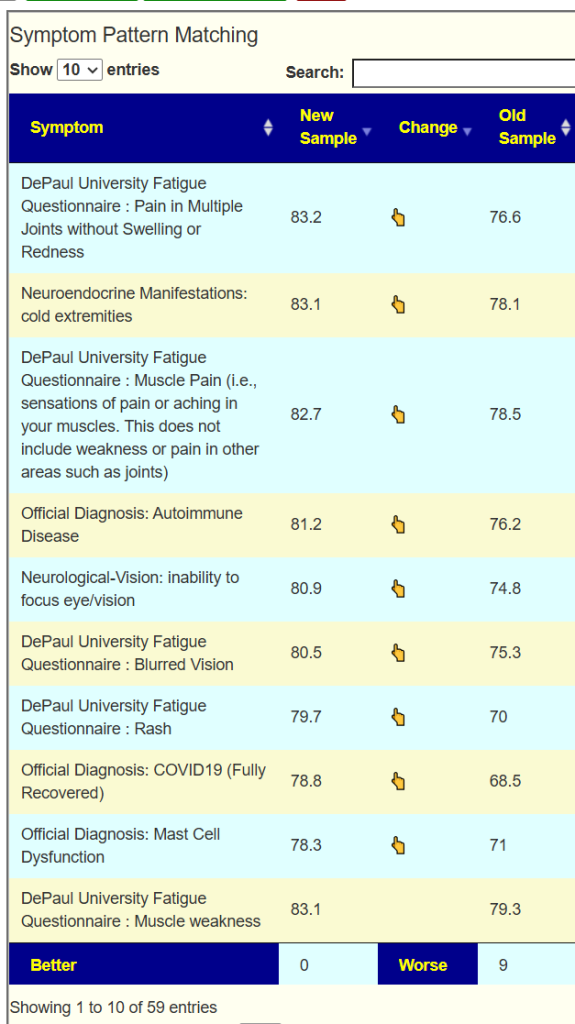
As often, we have a high hit rate of projected symptoms against actual symptoms.
Current Back Story
The first line is reflected above.
I have not been feeling so well lately (since the last year).
I would say that my symptoms has become worse.
Earlier it has always felt as I have done some progress but the last 12 months it has been the opposite.
Earlier I got rid of my muscle and joint pain but it has come back and I have much bigger issues with my red nose and my body feels very stressed.
Also feel very bloated.
A summary of my biggest issues:
- Get the red nose (some form of rosacea).
- Feel fatigued (both physically and mentally).
- Feeling stressed.
- Brain fog.
- Bloated.
- Lots of gas – I fart and burps a lot.
- Issues with allergies
- Muscle and joint pain
For the last 4 years I’ve been eating large amounts of rye and oats.
Around 150-200 gram of rye bread every day.
Around 70 gram of oats every day.
Been eating low fat, low protein and high carb (specially from rye, oats, apple juice and potatoes) because this diet seem to reduce my symptoms.
As soon as I start to eat high meat and high fat my symptoms get worse.
Traditional Analysis
First, I am doing the “traditional” analysis before exploring some work in progress to improve suggestions further. The process is simple, pick Beginner-Symptoms, mark symptoms and get suggestions. This is the process that seems to produce the best results. Other choices are intended to satisfy people with different assumptions. The site purpose is allow people to use the data according to their beliefs about the best way.
Despite having 42 symptoms entered, this boiled down to just 20 bacteria. Many related symptoms are connected to the same bacteria.
Investigate: “As soon as I start to eat high meat and high fat my symptoms”
Looking at the consensus report we see:
- high red meat: -155
- Pork : -22
- Chicken : -14
Which agrees with his reported response. On the other side, generic “fat” is a significant plus– so the type of fat seems to be critical.
Investigate Current Eating Habits: eating large amounts of rye and oats.
The suggestions are intended to be course corrections for the microbiome. Keeping on a course for too long may end up running aground on mudflats (instead of the original reefs that the course correction was intended for).
- Avena sativa {Oats} is a -16
- Secale cereale {Rye} +276
- Hordeum vulgare {Barley} -225
- Avena sativa x Hordeum vulgare {barley,oat} -240
- ß-glucan {Beta-Glucan} -110
- wheat, amaranth,quinoa, taro,buckwheat were all negatives
The question is why rye is ok and other grains are not? It may be due to some composition aspect or a side-effect of having sparse data. It looks like some change of diet is suggested.
The positive food items appear to be:
- fruit: grapes, especially lingonberries, cranberries (and likely cloudberries!), citric fruits, bananas
- fish: fish oil,
- legumes: beets, nuts, Asparagus, Rhubarb but NOT Pulses, Beans
- Quinoa – which unlike cereals above is gluten free
Vitamins
The list is pretty short.
- To take: Vitamin B6,7 and 12 but no other B vitamins (i.e. no mixtures)
- Iron and Vitamin D are fine
Probiotics
There are two approaches here.
- Using PubMed Studies only (sparse data, negative impacts almost never published)
- Using Inference from Microbiome Taxa R2 Site.
- A third way is in the next section with is like the R2 site, but using a 16s Biomesight reference set instead of a shotgun.
PubMed Studies
The top ones, in descending order are (with strike thru on ones that are hard to obtain):
Latilactobacillus sakei {Lactobacillus sakei}- Lacticaseibacillus paracasei {L.paracasei}
- Enterococcus faecium sf 68 {bioflorin}
- Escherichia coli Nissle 1917 {Mutaflor} os Symbioflor-2
Lentilactobacillus buchneri {Lactobacillus buchneri}- Lactococcus lactis {Streptococcus lactis}
- bifidobacterium bifidum {B. bifidum}
- aspergillus oryzae {koji}
Bacillus amyloliquefaciens group {B. Amyloliquefaciens}
Microbiome Taxa R2 Site
We end up with just 2 “safe” (positive impact only)
This leaves only one with 100% consensus: E.Coli (Mutaflor, Symbioflor-2). Looking at PubMed with a net positive with R2, we also have
My Own Experience
While fighting ME/CFS, I retested about 6 weeks after getting the results of the last test. I noticed that suggestions swing back and worth a lot – but I kept following them. Often there can be a battle between “common sense beliefs” and what the algorithms find. Avoiding something during one cycle and then taking it the next cycle seemed “irrational”. I borrowed from physical processes the concept of “microbiome oscillations” and stopped worrying about the swings.
My personal advice is simple, get results and then do suggestions for 6-8 week and do another test. With test processing delays, it means about 10-12 weeks on each set of suggestions
Going Forward — and a new Algorithm
Recently I have been working on an Odds Ratio investigation. The reason is simple, the Odds ratio gives an objective measure of the importance of each bacteria for the symptoms. The new approach uses Odds Ratio to determine the odds of a bacteria causing a symptom. The odds tells me the importance. If you are interested in more technical data, see:
I will be trying it out on his data. The databases involved are about 160GB with processing often taking 20 minutes for each processing state, so they are on my “garage” high performance server (nerd talk: 64GB of memory, fast M.2 NVMe 2TB drive for disk) and strictly for research/exploration at the moment.
Key differences
- we are going to estimate symptoms a different way than traditional (using odds)
- we are likely to have 100+ bacteria to shift
Predicted Symptoms Rank Order
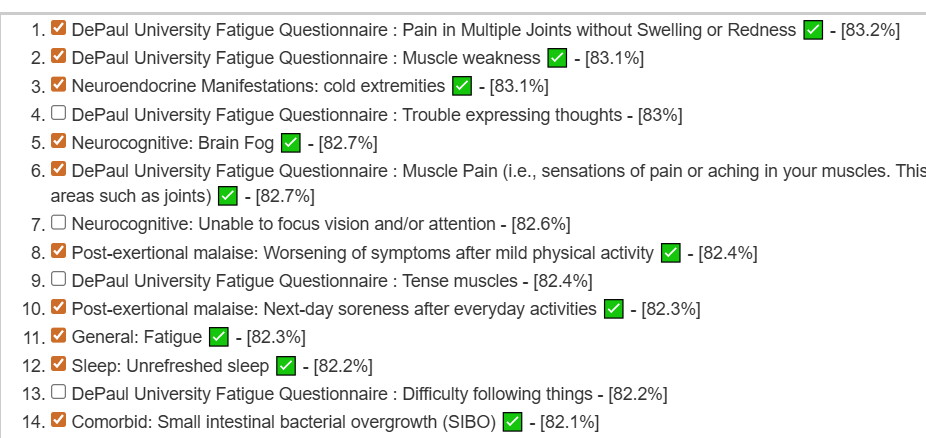
Using the Odds Ratio approach we get the following predictions that agrees with his reported symptoms/characteristics.
| Symptom Name | Strength |
| Age: 30-40 | 11.2 |
| Sleep: Unrefreshed sleep | 10.9 |
| Comorbid: Small intestinal bacterial overgrowth (SIBO) | 9.1 |
| Immune Manifestations: Inflammation (General) | 9 |
| General: Fatigue | 9 |
| DePaul University Fatigue Questionnaire : Tingling feeling | 8.8 |
| Neuroendocrine: Cold limbs (e.g. arms, legs hands) | 8.5 |
| Neuroendocrine Manifestations: cold extremities | 8.3 |
| Neurocognitive: Brain Fog | 8.1 |
| Post-exertional malaise: Worsening of symptoms after mild mental activity | 8 |
| DePaul University Fatigue Questionnaire : Fatigue | 7.9 |
| Gender: Male | 7.9 |
| DePaul University Fatigue Questionnaire : Muscle Pain (i.e., sensations of pain or aching in your muscles. This does not include weakness or pain in other areas such as joints) | 7.8 |
| Immune Manifestations: Bloating | 7.5 |
| DePaul University Fatigue Questionnaire : Allergies | 7.4 |
| DePaul University Fatigue Questionnaire : Muscle weakness | 6.9 |
| DePaul University Fatigue Questionnaire : Post-exertional malaise, feeling worse after doing activities that require either physical or mental exertion | 6.7 |
| DePaul University Fatigue Questionnaire : Rash | 6.5 |
| Post-exertional malaise: Mentally tired after the slightest effort | 6.3 |
| Comorbid: Histamine or Mast Cell issues | 6.3 |
| Post-exertional malaise: Next-day soreness after everyday activities | 6 |
| Post-exertional malaise: Muscle fatigue after mild physical activity | 6 |
| Official Diagnosis: Mast Cell Dysfunction | 5.9 |
| Neuroendocrine Manifestations: worsening of symptoms with stress. | 5.9 |
| Post-exertional malaise: Worsening of symptoms after mild physical activity | 5.8 |
| DePaul University Fatigue Questionnaire : Does physical activity make you feel worse | 5.7 |
| DePaul University Fatigue Questionnaire : Unrefreshing Sleep, that is waking up feeling tired | 5.7 |
| Immune: Flu-like symptoms | 5.7 |
| DePaul University Fatigue Questionnaire : Pain in Multiple Joints without Swelling or Redness | 5.5 |
| Neurological-Audio: hypersensitivity to noise | 5.2 |
| Immune Manifestations: Inflammation of skin, eyes or joints | 5.1 |
Looking at the existing estimates, we see far greater separation in weight/estimates. I favor separation because that implies much better focus on bacteria.
- DePaul University Fatigue Questionnaire : Pain in Multiple Joints without Swelling or Redness ✅ – [83.2%]
- DePaul University Fatigue Questionnaire : Muscle weakness ✅ – [83.1%]
- Neuroendocrine Manifestations: cold extremities ✅ – [83.1%]
- Neurocognitive: Brain Fog ✅ – [82.7%]
- DePaul University Fatigue Questionnaire : Muscle Pain (i.e., sensations of pain or aching in your muscles. This does not include weakness or pain in other areas such as joints) ✅ – [82.7%]
- Post-exertional malaise: Worsening of symptoms after mild physical activity ✅ – [82.4%]
- Post-exertional malaise: Next-day soreness after everyday activities ✅ – [82.3%]
- General: Fatigue ✅ – [82.3%]
- Sleep: Unrefreshed sleep ✅ – [82.2%]
- Comorbid: Small intestinal bacterial overgrowth (SIBO) ✅ – [82.1%]
- Immune Manifestations: Bloating ✅ – [81.9%]
- DePaul University Fatigue Questionnaire : Fatigue ✅ – [81.7%]
- Official Diagnosis: Chronic Fatigue Syndrome (CFS/ME) ✅ – [81.5%]
- Neurological-Audio: hypersensitivity to noise ✅ – [81.5%]
- DePaul University Fatigue Questionnaire : Unrefreshing Sleep, that is waking up feeling tired ✅ – [81.4%]
- DePaul University Fatigue Questionnaire : Difficulty staying asleep ✅ – [81.3%]
- Official Diagnosis: Autoimmune Disease ✅ – [81.2%]
- Immune Manifestations: Inflammation (General) ✅ – [81.2%]
- DePaul University Fatigue Questionnaire : Easily irritated – [81.2%]
- Neuroendocrine Manifestations: worsening of symptoms with stress. ✅ – [81.2%]
Key Bacteria identified
The new approach identifies these bacteria to target, with their relative importance (Weight). I just did another post on a ME/CFSer, Microbiome Interpretation – Questions From A User. Megamonas also was her top one.
| tax name | tax rank | Weight | Target |
| Megamonas | genus | 108.9 | Too High |
| Klebsiella oxytoca | species | 95.8 | Too High |
| Ruminococcus bromii | species | -17.9 | Too Low |
| Eubacteriales | order | -16.1 | Too Low |
| Clostridia | class | -15.1 | Too Low |
| Megamonas funiformis | species | 15 | Too High |
| Bacillota | phylum | -14.2 | Too Low |
| Ruminococcaceae | family | -14.2 | Too Low |
| Segatella | genus | 13.6 | Too High |
| Oscillospiraceae | family | -13.4 | Too Low |
| Ruminococcus | genus | -12.4 | Too Low |
| Terrabacteria group | clade | -12.4 | Too Low |
| Segatella copri | species | 11.9 | Too High |
| Bacteroidia | class | 11.4 | Too High |
| Bacteroidales | order | 11.4 | Too High |
| Bacteroidota/Chlorobiota group | clade | 10.3 | Too High |
| Bacteroidota | phylum | 10.1 | Too High |
| FCB group | clade | 9.8 | Too High |
| Prevotella | genus | 9.7 | Too High |
| Lachnospiraceae | family | -9.3 | Too Low |
| Prevotellaceae | family | 9 | Too High |
| Phocaeicola vulgatus | species | 8.4 | Too High |
| Akkermansiaceae | family | 6.7 | Too High |
| Bacteroides uniformis | species | 6 | Too High |
| Pseudomonadota | phylum | 5.9 | Too High |
| Gammaproteobacteria | class | 5.2 | Too High |
| Yersinia | genus | 4.8 | Too High |
| Verrucomicrobiota | phylum | 4.3 | Too High |
| Akkermansia | genus | 4.1 | Too High |
| Verrucomicrobiales | order | 4 | Too High |
The traditional approach identifies the list below. There is relatively little overlap. My ‘gut’ reading is that those above are likely a better candidate set than those below.
| Bacteria | Rank | Shift |
| Thiotrichales | order | High |
| Sharpea | genus | High |
| Selenomonas | genus | High |
| Ruminococcus | genus | High |
| Negativicutes | class | Low |
| Johnsonella | genus | High |
| Holdemania | genus | High |
| Erysipelothrix | genus | High |
| Dorea | genus | High |
| Desulfovibrionia | class | Low |
| delta/epsilon subdivisions | clade | Low |
| Cyanophyceae | class | Low |
| Cyanobacteriota/Melainabacteria group | clade | Low |
| Coprococcus | genus | High |
| Chlorobiota | phylum | High |
| Chlorobiia | class | High |
| Chlorobiaceae | family | High |
| Chlorobaculum | genus | High |
| Actinomycetota | phylum | Low |
| Actinomycetes | class | Low |
Suggestions
Since two bacteria dominates, I ran the suggestion algorithm only on those two bacteria. The results are below and very similar to the results from the traditional approach. “All algorithms lead to the same suggestions”. Doing the full list cited above, produced very similar suggestions.
| Modifier | Net |
| fruit/legume fibre | 263 |
| fruit | 241 |
| Fiber, total dietary | 217 |
| Chitosan | 217 |
| Slow digestible carbohydrates. {Low Glycemic} | 210 |
| oolong teas | 205 |
| polyphenols | 203 |
| resveratrol-pterostilbene x Quercetin {quercetin x resveratrol} | 202 |
| (2->1)-beta-D-fructofuranan {Inulin} | 201 |
| 3,3′,4′,5,7-pentahydroxyflavone {Quercetin} | 200 |
| High-fibre diet {Whole food diet} | 199 |
| Citrus limon {Lemon} | 194 |
| 5,6-dihydro-9,10-dimethoxybenzo[g]-1,3-benzodioxolo[5,6-a]quinolizinium {Berberine} | 193 |
| Lonicera periclymenum {Epazote} | 184 |
| Grape Polyphenols {Grape Flavonoids} | 177 |
| tea | 177 |
| dietary fiber | 169 |
| red wine | 168 |
| grapes | 163 |
| Linum usitatissimum {Flaxseed} | 160 |
| Lactobacillus plantarum {L. plantarum} | 158 |
| Camellia Linnaeus {camellia} | 158 |
| Coffee | 150 |
| Hydrastis canadensis {Goldenseal} | 148 |
| Ethanoic acid {Vinegar} | 148 |
| Ulmus rubra {slippery elm} | 145 |
| Ligilactobacillus salivarius {L. salivarius} | 144 |
| Ginkgo biloba {Ginkgo} | 139 |
| Cola aspartame {Diet Cola} | 138 |
| pseudo-cereals {amaranth,quinoa, taro,buckwheat} | 137 |
| Agaricus bisporus {White button mushrooms} | 134 |
| Theobroma cacao {Cacao} | 131 |
| Caffeine | 127 |
| Lacticaseibacillus rhamnosus {l. rhamnosus} | 124 |
| fucoidan {Brown Algae Extract} | 122 |
| Litchi chinensis {lychee fruit} | 121 |
| coptis chinensis {Chinese goldthread } | 121 |
Probiotics using R2
First I used only the top two bacteria to see what is suggested with a very targeted set.
- Bacillus subtilis 75
- Lactobacillus jensenii 60
Below are pushing the full set of identified bacteria through BiomeSight R2 matrix, then filtered to positive impact with no risk. Escherichia coli (cited above) continues to be a take. Bacillus subtilis would be my fall back suggestion for a probiotic. It is marginally negative on the consensus report and not cited on other R2 suggestion list. For others candidates
- Lactobacillus jensenii was one for and no comment
- Lactococcus lactis is a one for and one against
- Lactobacillus helveticus is one strong against and no comment
- Lacticaseibacillus casei is one strong against and no comment
- Akkermansia muciniphila is two strong avoid
I tend to do a variation of the traditional “Do no harm”, minimize the risk of adverse shifts.
| Probiotic | Net Impact | Good Count | Bad Count |
| Bacillus subtilis | 81.9 | 5 | 0 |
| Lactobacillus jensenii | 68.5 | 2 | 0 |
| Lactococcus lactis | 26.8 | 2 | 0 |
| Lactobacillus helveticus | 21.4 | 4 | 0 |
| Lacticaseibacillus casei | 21.4 | 2 | 0 |
| Segatella copri | 21.2 | 3 | 0 |
| Lactiplantibacillus pentosus | 21 | 5 | 0 |
| Akkermansia muciniphila | 18.7 | 4 | 0 |
| Bacillus amyloliquefaciens group | 18.3 | 1 | 0 |
| Enterococcus faecium | 16.3 | 1 | 0 |
| Limosilactobacillus fermentum | 13.6 | 1 | 0 |
| Heyndrickxia coagulans | 13.4 | 3 | 0 |
| Enterococcus durans | 12.4 | 2 | 0 |
| Pediococcus acidilactici | 11 | 1 | 0 |
| Enterococcus faecalis | 10.6 | 2 | 0 |
| Leuconostoc mesenteroides | 10.5 | 1 | 0 |
| Escherichia coli | 4.3 | 3 | 0 |
Bottom Line
The purpose of this post was to evaluate suggestions for a regular reader. The secondary goal was to see how well a new approach that I am developing is working. This new approach produces different targeted bacteria with very similar suggestions generated, the most significance difference is far more targeted probiotics for the symptoms based on the same lab data.
The one interesting aspect is that the key bacteria (just 2) were clearly identified. These two bacteria alone produced suggestions similar to the bigger bacteria selection. I do like this narrow bacteria selection of key bacteria and will likely do a few more samples to further explore things.
Follow Up
I decided to look at all of his samples with the new algorithm to look for patterns. Megamonas stands out as the one that most frequently appears and disappears. Klebsiella oxytoca and Morganellaceae are the next candidates.
| Upload Date | Top Bacteria |
| 2021-09-24 | Megamonas genus 108.9 Too High Lachnospiraceae family -56.2 Too Low Eubacteriales order -47.3 Too Low Bacillota phylum -46.6 Too Low Clostridia class -46.4 Too Low Terrabacteria group clade -44.8 Too Low |
| 2021-09-24 | Bacillota phylum -46.5 Too Low Eubacteriales order -45.1 Too Low Terrabacteria group clade -45 Too Low Clostridia class -44.1 Too Low Lachnospiraceae family -43.6 Too Low |
| 2022-04-19 | Klebsiella oxytoca species 95.8 Too High Morganellaceae family 91 Too High |
| 2022-09-04 | Oscillospiraceae family 17.7 Too High Ruminococcaceae family 17.4 Too High Bacteroidaceae family -16 Too Low Bacteroides genus -15.9 Too Low |
| 2023-03-15 | Megamonas genus 108.9 Too High Lachnospiraceae family -26 Too Low |
| 2023-09-26 | Megamonas genus 108.9 Too High |
| 2024-02-13 | Bacteroidaceae family -27.3 Too Low Bacteroides genus -27.3 Too Low Phocaeicola dorei species -21.4 Too Low |
| 2024-09-25 | Segatella genus 13.6 Too High Segatella copri species 11.9 Too High |
| 2025-04-22 | Morganellaceae family 91.6 Too High |
| 2025-12-08 | Megamonas genus 108.9 Too High Klebsiella oxytoca species 95.8 Too High |
Recent Comments