Microbiome Prescription has a rich collection of annotated samples from different labs (uBiome, Ombre, Biomesight). The samples are annotated with self declared symptoms from a list of 548 different symptoms/diagnosis. 328 symptoms had statistically significant associations.
- Biomesight: 4169 samples
- Ombre: 1514 samples
- uBiome: 795 samples
There are several possibility of associations to these symptoms, including:
- Bacteria Association
- Enzyme Association
- Metabolite Association which we can decompose into
- Production
- Substrate (Consumers)
- Net Metabolite (Production – Consumer)
For each of these 5 vectors, we use these three statistical methods and set out criteria to p < 0.005:
- Fisher’s exact test on prevalence of bacteria
- Mann Whitney Wilcoxon Test
- t-Test on Means
We used KEGG.JP data as a poor man method of compute metabolites.
To go directly to the page reporting data: Symptom Association
This processing took a few days hammering a PC at 90% CPU.
https://www.youtube.com/watch?v=sM4V5hesb1s
High Level Overview
Below we have counts of the associations found. It is clear that bacteria associations are weaker(fewer) than Enzymes by a factor of 4-10. With metabolites, the net metabolite appears a poorer estimator than either producers or substrates.
As would be expected, large population, we find more associations as the population increases.
Bacteria
| Source | Associations |
| BiomeSight | 33432 |
| Thryve | 28668 |
| uBiome | 9283 |
Enzymes
| Source | Associations |
| BiomeSight | 325170 |
| Thryve | 78485 |
| uBiome | 41707 |
Metabolite Production
| Source | Associations |
| BiomeSight | 226447 |
| Thryve | 49122 |
| uBiome | 26756 |
Metabolite Substrate
| Source | Associations |
| BiomeSight | 227053 |
| Thryve | 50164 |
| uBiome | 28162 |
Metabolite Net
| Source | Associations |
| BiomeSight | 123631 |
| Thryve | 25493 |
| uBiome | 14771 |
Data Accessibility
The findings are available on microbiome prescription

The key thing to be aware of is that the results are different using data from each lab. For background see Nightmare.

The page is simple:
- Pick the Symptoms
- Pick the number of TOP items you want from each lab.

This is then followed by 5 sortable and filterable tables.
Examples of agreement
For ME/CFS we have all three labs reporting these bacteria are significant:
- Porphyromonas genus
- Bifidobacterium genus
- Bifidobacteriaceae family
- Bifidobacteriales order
- Hungateiclostridiaceae family
For Enzymes:
- 1.1.1.65 pyridoxine 4-dehydrogenase
- 1.8.98.6 formate:CoB-CoM heterodisulfide,ferredoxin reductase
- 2.1.1.172 16S rRNA (guanine1207–N2)-methyltransferase
- 2.3.2.5 glutaminyl-peptide cyclotransferase
- 2.4.1.336 monoglucosyldiacylglycerol synthase
- 2.6.1.59 dTDP-4-amino-4,6-dideoxygalactose transaminase
- 2.7.7.15 choline-phosphate cytidylyltransferase
- 2.7.8.36 undecaprenyl phosphate N,N′-diacetylbacillosamine 1-phosphate transferase
- 3.4.21.83 oligopeptidase B
- 4.1.1.31 phosphoenolpyruvate carboxylase
For Net Metabolite we have a much longer list:
- C01142 (3S)-3,6-Diaminohexanoate
- C20748 (E)-4-(Trimethylammonio)but-2-enoyl-CoA
- C02612 (R)-2-Methylmalate
- C06010 (S)-2-Acetolactate
- C00424 (S)-Lactaldehyde
- C07281 [eIF5A-precursor]-lysine
- C15811 [Enzyme]-cysteine
- C15812 [Enzyme]-S-sulfanylcysteine
- C16236 [Protein]-N6-(octanoyl)-L-lysine
- C16832 [Protein]-N6-[(R)-dihydrolipoyl]-L-lysine
- C21440 [Protein]-S-sulfanyl-L-cysteine
- C01302 1-(2-Carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate
- C04751 1-(5-Phospho-D-ribosyl)-5-amino-4-imidazolecarboxylate
- C04677 1-(5′-Phosphoribosyl)-5-amino-4-imidazolecarboxamide
- C06364 1,2-Diacyl-3-alpha-D-glucosyl-sn-glycerol
- C00641 1,2-Diacyl-sn-glycerol
- C15606 1,2-Dihydroxy-5-(methylthio)pent-1-en-3-one
- C03657 1,4-Dihydroxy-2-naphthoate
- C11437 1-Deoxy-D-xylulose 5-phosphate
- C04006 1D-myo-Inositol 3-phosphate
- C11811 1-Hydroxy-2-methyl-2-butenyl 4-diphosphate
- C03972 2,3,4,5-Tetrahydrodipicolinate
- C00691 2,4,6/3,5-Pentahydroxycyclohexanone
- C21607 2,6-Di-O-alpha-D-mannosyl-1-phosphatidyl-1D-myo-inositol
- C04691 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate
- C20905 2-Iminobutanoate
- C20904 2-Iminopropanoate
- C21609 2-O-(6-O-Acyl-alpha-D-mannosyl)-1-phosphatidyl-1D-myo-inositol
- C05807 2-Polyprenylphenol
- C16463 3′,5′-Cyclic diGMP
- C20934 3-Deoxy-D-glycero-D-galacto-non-2-ulopyranosonate
- C04478 3-Deoxy-D-manno-octulosonate 8-phosphate
- C00587 3-Hydroxybenzoate
- C00141 3-Methyl-2-oxobutanoic acid
- C00053 3′-Phosphoadenylyl sulfate
- C00197 3-Phospho-D-glycerate
- C11435 4-(Cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- C11355 4-Amino-4-deoxychorismate
- C04752 4-Amino-5-hydroxymethyl-2-methylpyrimidine diphosphate
- C00568 4-Aminobenzoate
- C00334 4-Aminobutanoate
- C04327 4-Methyl-5-(2-phosphooxyethyl)thiazole
- C01180 4-Methylthio-2-oxobutanoic acid
- C22411 4-O-{Poly[(2R)-glycerophospho]-(2R)-glycerophospho}-N-acetyl-beta-D-mannosaminyl-(1->4)-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
- C21502 4-O-Di[(2R)-1-glycerophospho]-N-acetyl-beta-D-mannosaminyl-(1->4)-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
- C03393 4-Phospho-D-erythronate
- C00445 5,10-Methenyltetrahydrofolate
- C00143 5,10-Methylenetetrahydrofolate
- C03089 5-Methylthio-D-ribose
- C00119 5-Phospho-alpha-D-ribose 1-diphosphate
- C01300 6-(Hydroxymethyl)-7,8-dihydropterin
- C01019 6-Deoxy-L-galactose
- C04807 6-Hydroxymethyl-7,8-dihydropterin diphosphate
- C20176 8-Oxo-dGDP
- C22235 8-Oxo-GDP
- C22382 Aceneuramate
- C00033 Acetate
- C00227 Acetyl phosphate
- C00024 Acetyl-CoA
- C00147 Adenine
- C00212 Adenosine
- C06508 Adenosyl cobinamide
- C06506 Adenosyl cobyrinate a,c diamide
- C00008 ADP
- C00179 Agmatine
- C00069 Alcohol
- C00267 alpha-D-Glucose
- C20237 alpha-Maltose 1-phosphate
- C00014 Ammonia
- C00020 AMP
- C00002 ATP
- C05345 beta-D-Fructose 6-phosphate
- C00663 beta-D-Glucose 1-phosphate
- C00576 Betaine aldehyde
- C20568 beta-L-Arabinofuranosyl-(1->2)-beta-L-arabinofuranose
- C00120 Biotin
- C00323 Caffeoyl-CoA
- C00169 Carbamoyl phosphate
- C00090 Catechol
- C00307 CDP-choline
- C00269 CDP-diacylglycerol
- C00513 CDP-glycerol
- C00114 Choline
- C00588 Choline phosphate
- C00011 CO2
- C00010 CoA
- C06504 Cob(II)yrinate a,c diamide
- C11545 Cobalt-precorrin 8
- C05773 Cobyrinate
- C00876 Coenzyme F420
- C00063 CTP
- C03492 D-4′-Phosphopantothenate
- C00405 D-Amino acid
- C00239 dCMP
- C17010 Dehypoxanthine futalosine
- C02269 Deoxynucleoside
- C00677 Deoxynucleoside triphosphate
- C04666 D-erythro-1-(Imidazol-4-yl)glycerol 3-phosphate
- C00095 D-Fructose
- C00354 D-Fructose 1,6-bisphosphate
- C00333 D-Galacturonate
- C00257 D-Gluconic acid
- C00198 D-Glucono-1,5-lactone
- C00329 D-Glucosamine
- C00031 D-Glucose
- C00103 D-Glucose 1-phosphate
- C00258 D-Glycerate
- C00235 Dimethylallyl diphosphate
- C00013 Diphosphate
- C17556 di-trans,poly-cis-Undecaprenyl phosphate
- C00159 D-Mannose
- C00636 D-Mannose 1-phosphate
- C00039 DNA
- C00110 Dolichyl phosphate
- C03862 Dolichyl phosphate D-mannose
- C00121 D-Ribose
- C01151 D-Ribose 1,5-bisphosphate
- C03319 dTDP-L-rhamnose
- C00365 dUMP
- C00460 dUTP
- C00231 D-Xylulose 5-phosphate
- C00125 Ferricytochrome c
- C00126 Ferrocytochrome c
- C00406 Feruloyl-CoA
- C00061 FMN
- C00122 Fumarate
- C16999 Futalosine
- C02686 Galactosylceramide
- C00096 GDP-mannose
- C00353 Geranylgeranyl diphosphate
- C00051 Glutathione
- C00116 Glycerol
- C00184 Glycerone
- C00111 Glycerone phosphate
- C00037 Glycine
- C00266 Glycolaldehyde
- C00160 Glycolate
- C02412 Glycyl-tRNA(Gly)
- C00048 Glyoxylate
- C00044 GTP
- C00080 H+
- C00288 HCO3-
- C06250 Holo-[carboxylase]
- C04298 Holo-[citrate (pro-3S)-lyase]
- C00283 Hydrogen sulfide
- C06399 Hydrogenobyrinate
- C00530 Hydroquinone
- C15603 Hydroquinone
- C15809 Iminoglycine
- C00130 IMP
- C00463 Indole
- C03506 Indoleglycerol phosphate
- C00885 Isochorismate
- C00129 Isopentenyl diphosphate
- C03508 L-2-Amino-3-oxobutanoic acid
- C15556 L-3,4-Dihydroxybutan-2-one 4-phosphate
- C00041 L-Alanine
- C00886 L-Alanyl-tRNA
- C00049 L-Aspartate
- C00441 L-Aspartate 4-semialdehyde
- C20750 L-Carnitinyl-CoA
- C00327 L-Citrulline
- C02291 L-Cystathionine
- C00097 L-Cysteine
- C02700 L-Formylkynurenine
- C00025 L-Glutamate
- C01165 L-Glutamate 5-semialdehyde
- C00064 L-Glutamine
- C02047 L-Leucyl-tRNA
- C00073 L-Methionine
- C00077 L-Ornithine
- C00508 L-Ribulose
- C00065 L-Serine
- C00188 L-Threonine
- C00083 Malonyl-CoA
- C05819 Menaquinol
- C00409 Methanethiol
- C06717 Mycothiol
- C00137 myo-Inositol
- C04916 N-(5′-Phospho-D-1′-ribulosylformimino)-5-amino-1-(5”-phospho-D-ribosyl)-4-imidazolecarboxamide
- C20390 N-Acetyl-alpha-D-glucosaminyl-diphospho-trans,octacis-decaprenol
- C00140 N-Acetyl-D-glucosamine
- C00003 NAD+
- C00004 NADH
- C00006 NADP+
- C00005 NADPH
- C00436 N-Carbamoylputrescine
- C00153 Nicotinamide
- C00201 Nucleoside triphosphate
- C00979 O-Acetyl-L-serine
- C00009 Orthophosphate
- C22151 Oxidized [2Fe-2S] ferredoxin
- C01134 Pantetheine 4′-phosphate
- C00864 Pantothenate
- C00472 p-Benzoquinone
- C00416 Phosphatidate
- C03167 Phosphonoacetaldehyde
- C19692 Polysulfide
- C06408 Precorrin 8X
- C00585 Protein tyrosine
- C01167 Protein tyrosine phosphate
- C02880 Protochlorophyllide
- C00134 Putrescine
- C00250 Pyridoxal
- C00022 Pyruvate
- C15602 Quinone
- C22150 Reduced [2Fe-2S] ferredoxin
- C01080 Reduced coenzyme F420
- C00473 Retinol
- C00255 Riboflavin
- C00046 RNA
- C00021 S-Adenosyl-L-homocysteine
- C00019 S-Adenosyl-L-methionine
- C04188 S-Methyl-5-thio-D-ribose 1-phosphate
- C04582 S-Methyl-5-thio-D-ribulose 1-phosphate
- C00623 sn-Glycerol 1-phosphate
- C00315 Spermidine
- C03539 S-Ribosyl-L-homocysteine
- C00059 Sulfate
- C00704 Superoxide
- C00378 Thiamine
- C00422 Triacylglycerol
- C00066 tRNA
- C01642 tRNA(Gly)
- C01645 tRNA(Leu)
- C00015 UDP
- C19725 UDP-2,3-diacetamido-2,3-dideoxy-alpha-D-glucuronate
- C00043 UDP-N-acetyl-alpha-D-glucosamine
- C00203 UDP-N-acetyl-D-galactosamine
- C01050 UDP-N-acetylmuramate
- C05892 UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine
- C00105 UMP
- C00106 Uracil
- C01051 Uroporphyrinogen III
- C00001 Water
- C00385 Xanthine
- C00655 Xanthosine 5′-phosphate
For Metabolite Producers:
- C22302 (+)-6-Hydroxypinoresinol
- C19831 (1R,6S)-6-Amino-5-oxocyclohex-2-ene-1-carboxylate
- C05161 (2R,5S)-2,5-Diaminohexanoate
- C04236 (2S)-2-Isopropyl-3-oxosuccinate
- C20258 (2S,4S)-4-Hydroxy-2,3,4,5-tetrahydrodipicolinate
- C01142 (3S)-3,6-Diaminohexanoate
- C00566 (3S)-Citryl-CoA
- C01186 (3S,5S)-3,5-Diaminohexanoate
- C00712 (9Z)-Octadecenoic acid
- C20748 (E)-4-(Trimethylammonio)but-2-enoyl-CoA
- C22385 (L-Cysteinyl)adenylate
- C02489 (R)-2-Hydroxyacid
- C02612 (R)-2-Methylmalate
- C04352 (R)-4′-Phosphopantothenoyl-L-cysteine
- C00810 (R)-Acetoin
- C00937 (R)-Lactaldehyde
- C03912 (S)-1-Pyrroline-5-carboxylate
- C06010 (S)-2-Acetolactate
- C11838 (S)-4,5-Dihydroxypentane-2,3-dione
- C03656 (S)-5-Amino-3-oxohexanoic acid
- C00424 (S)-Lactaldehyde
- C21068 [5-(Aminomethyl)furan-3-yl]methyl phosphate
- C15977 [Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase] S-(2-methylpropanoyl)dihydrolipoyllysine
- C16254 [Dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine
- C07282 [eIF5A-precursor]-deoxyhypusine
- C07281 [eIF5A-precursor]-lysine
- C15811 [Enzyme]-cysteine
- C15812 [Enzyme]-S-sulfanylcysteine
- C22155 [Fe-S] cluster scaffold protein
- C01281 [L-Glutamate:ammonia ligase (ADP-forming)]
- C20730 [Protein]-FMN-L-Threonine
- C16236 [Protein]-N6-(octanoyl)-L-lysine
- C16832 [Protein]-N6-[(R)-dihydrolipoyl]-L-lysine
- C01242 [Protein]-S8-aminomethyldihydrolipoyllysine
- C21440 [Protein]-S-sulfanyl-L-cysteine
- C21879 1-(5-O-Phospho-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium-3-carbonyl adenylate
- C04751 1-(5-Phospho-D-ribosyl)-5-amino-4-imidazolecarboxylate
- C04823 1-(5′-Phosphoribosyl)-5-amino-4-(N-succinocarboxamide)-imidazole
- C04677 1-(5′-Phosphoribosyl)-5-amino-4-imidazolecarboxamide
- C04734 1-(5′-Phosphoribosyl)-5-formamido-4-imidazolecarboxamide
- C02490 1,2-beta-D-Glucan
- C00641 1,2-Diacyl-sn-glycerol
- C00986 1,3-Diaminopropane
- C06485 1,5-Anhydro-D-fructose
- C00234 10-Formyltetrahydrofolate
- C21835 12,18-Didecarboxysiroheme
- C04230 1-Acyl-sn-glycero-3-phosphocholine
- C11437 1-Deoxy-D-xylulose 5-phosphate
- C04006 1D-myo-Inositol 3-phosphate
- C11557 1-Phosphatidyl-1D-myo-inositol 5-phosphate
- C19771 2′-(5-Triphosphoribosyl)-3′-dephospho-CoA
- C04640 2-(Formamido)-N1-(5′-phosphoribosyl)acetamidine
- C01159 2,3-Bisphospho-D-glycerate
- C00691 2,4,6/3,5-Pentahydroxycyclohexanone
- C20424 2,4-Diacetamido-2,4,6-trideoxy-D-mannopyranose
- C21896 2,4-Diacetylphloroglucinol
- C02780 2,5-Didehydro-D-gluconate
- C21607 2,6-Di-O-alpha-D-mannosyl-1-phosphatidyl-1D-myo-inositol
- C00900 2-Acetolactate
- C17234 2-Aminobut-2-enoate
- C11453 2-C-Methyl-D-erythritol 2,4-cyclodiphosphate
- C04442 2-Dehydro-3-deoxy-6-phospho-D-gluconate
- C04691 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate
- C03827 2-Dehydro-3-deoxy-L-fuconate
- C06892 2-Deoxy-5-keto-D-gluconic acid
- C00672 2-Deoxy-D-ribose 1-phosphate
- C06196 2′-Deoxyinosine 5′-phosphate
- C19970 2-Hydroxy-dAMP
- C20905 2-Iminobutanoate
- C02631 2-Isopropylmaleate
- C02222 2-Maleylacetate
- C02226 2-Methylmaleate
- C21608 2-O-(6-O-Acyl-alpha-D-mannosyl)-6-O-alpha-D-mannosyl-1-phosphatidyl-1D-myo-inositol
- C19792 2-O-(alpha-D-Glucopyranosyl)-D-glycerate
- C03586 2-Oxo-2,3-dihydrofuran-5-acetate
- C06054 2-Oxo-3-hydroxy-4-phosphobutanoate
- C00109 2-Oxobutanoate
- C00026 2-Oxoglutarate
- C11436 2-Phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- C00631 2-Phospho-D-glycerate
- C13309 2-Phytyl-1,4-naphthoquinone
- C05807 2-Polyprenylphenol
- C01267 3-(Imidazol-4-yl)-2-oxopropyl phosphate
- C00575 3′,5′-Cyclic AMP
- C16463 3′,5′-Cyclic diGMP
- C20772 3-[(1-Carboxyvinyl)oxy]benzoate
- C00944 3-Dehydroquinate
- C02637 3-Dehydroshikimate
- C20934 3-Deoxy-D-glycero-D-galacto-non-2-ulopyranosonate
- C21383 3-Deoxy-D-glycero-D-galacto-non-2-ulopyranosonate 9-phosphate
- C04046 3-D-Glucosyl-1,2-diacylglycerol
- C20960 3-Hydroxy-5-phosphooxypentane-2,4-dione
- C00141 3-Methyl-2-oxobutanoic acid
- C03069 3-Methylcrotonyl-CoA
- C00685 3-Oxoacyl-[acyl-carrier protein]
- C02941 3-Oxo-Delta1-steroid
- C00619 3-Oxo-Delta4-steroid
- C00053 3′-Phosphoadenylyl sulfate
- C00197 3-Phospho-D-glycerate
- C00236 3-Phospho-D-glyceroyl phosphate
- C03232 3-Phosphonooxypyruvate
- C02798 3-Phosphonopyruvate
- C22313 3-Sulfinopropanoate
- C11435 4-(Cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- C16144 4,4′-Diapophytoene
- C04556 4-Amino-2-methyl-5-(phosphooxymethyl)pyrimidine
- C01279 4-Amino-5-hydroxymethyl-2-methylpyrimidine
- C04752 4-Amino-5-hydroxymethyl-2-methylpyrimidine diphosphate
- C00334 4-Aminobutanoate
- C05848 4-Hydroxy-3-polyprenylbenzoate
- C00156 4-Hydroxybenzoate
- C00233 4-Methyl-2-oxopentanoate
- C04327 4-Methyl-5-(2-phosphooxyethyl)thiazole
- C22411 4-O-{Poly[(2R)-glycerophospho]-(2R)-glycerophospho}-N-acetyl-beta-D-mannosaminyl-(1->4)-N-acetyl-alpha-D-glucosaminyl-diphospho-ditrans,octacis-undecaprenol
- C02964 4-O-beta-D-Glucopyranosyl-D-mannose
- C19877 4-O-Phospho-alpha-Kdo-(2->6)-lipid IVA
- C03082 4-Phospho-L-aspartate
- C04294 5-(2-Hydroxyethyl)-4-methylthiazole
- C04896 5-(5-Phospho-D-ribosylaminoformimino)-1-(5-phosphoribosyl)-imidazole-4-carboxamide
- C00445 5,10-Methenyltetrahydrofolate
- C00431 5-Aminopentanoate
- C21877 5-Carboxy-1-(5-O-phospho-beta-D-ribofuranosyl)pyridin-1-ium-3-carbonyl adenylate
- C15667 5-Carboxyamino-1-(5-phospho-D-ribosyl)imidazole
- C16737 5-Deoxy-D-glucuronate
- C22288 5-Deoxy-D-ribose
- C03089 5-Methylthio-D-ribose
- C01269 5-O-(1-Carboxyvinyl)-3-phosphoshikimate
- C02805 5-Oxoprolyl-peptide
- C00119 5-Phospho-alpha-D-ribose 1-diphosphate
- C03838 5′-Phosphoribosylglycinamide
- C19787 5′-S-Methyl-5′-thioinosine
- C03773 6-Acetyl-beta-D-galactoside
- C20773 6-Amino-6-deoxyfutalosine
- C02954 6-Aminopenicillanate
- C20830 6-Deoxy-6-sulfo-D-fructose
- C19859 6-Methoxy-3-methyl-2-all-trans-polyprenyl-1,4-benzoquinol
- C00345 6-Phospho-D-gluconate
- C01037 7,8-Diaminononanoate
- C15858 7,9,7′,9′-tetracis-Lycopene
- C19759 7,9,9′-tricis-Neurosporene
- C04643 7-Oxodeoxycholate
- C20176 8-Oxo-dGDP
- C19968 8-Oxo-dGMP
- C22235 8-Oxo-GDP
- C14909 9alpha-Hydroxyandrosta-1,4-diene-3,17-dione
- C00028 Acceptor
- C00033 Acetate
- C05744 Acetoacetyl-[acp]
- C00207 Acetone
- C00227 Acetyl phosphate
- C00024 Acetyl-CoA
- C00147 Adenine
- C00212 Adenosine
- C06507 Adenosyl cobyrinate hexaamide
- C01299 Adenylyl-[L-glutamate:ammonia ligase (ADP-forming)]
- C00008 ADP
- C20784 ADP-5-ethyl-4-methylthiazole-2-carboxylate
- C00498 ADP-glucose
- C00499 Allantoate
- C04145 all-trans-Nonaprenyl diphosphate
- C04146 all-trans-Octaprenyl diphosphate
- C04465 alpha,alpha’-Trehalose 6,6′-bismycolate
- C00446 alpha-D-Galactose 1-phosphate
- C00936 alpha-D-Mannose
- C02504 alpha-Isopropylmalate
- C06365 alpha-Kojibiosyldiacylglycerol
- C01888 Aminoacetone
- C03373 Aminoimidazole ribotide
- C00014 Ammonia
- C00020 AMP
- C06697 Arsenite
- C00002 ATP
- C21336 beta-1,2-Mannobiose
- C05345 beta-D-Fructose 6-phosphate
- C00221 beta-D-Glucose
- C00663 beta-D-Glucose 1-phosphate
- C01172 beta-D-Glucose 6-phosphate
- C20569 beta-L-Arabinofuranose
- C01563 Carbamate
- C00169 Carbamoyl phosphate
- C04419 Carboxybiotin-carboxyl-carrier protein
- C20969 Carboxyphosphate
- C05980 Cardiolipin
- C00307 CDP-choline
- C00269 CDP-diacylglycerol
- C00789 CDP-ribitol
- C05306 Chlorophyll a
- C00114 Choline
- C01794 Choloyl-CoA
- C00251 Chorismate
- C00417 cis-Aconitate
- C00158 Citrate
- C20419 CMP-N,N’-diacetyllegionaminate
- C00011 CO2
- C00010 CoA
- C00175 Cobalt ion
- C16242 Cobalt-precorrin 5A
- C11545 Cobalt-precorrin 8
- C05773 Cobyrinate
- C19724 Cobyrinate c-monamide
- C04832 Coenzyme M 7-mercaptoheptanoylthreonine-phosphate heterodisulfide
- C03263 Coproporphyrinogen III
- C00063 CTP
- C00475 Cytidine
- C22216 Cytidine 5′-{[hydroxy(2-hydroxyethyl)phosphonoyl]phosphate}
- C04122 D-1-Aminopropan-2-ol O-phosphate
- C03492 D-4′-Phosphopantothenate
- C00993 D-Alanyl-D-alanine
- C00405 D-Amino acid
- C00360 dAMP
- C00705 dCDP
- C00239 dCMP
- C03112 Deacetylcephalosporin C
- C00857 Deamino-NAD+
- C15495 Decylubiquinol
- C15853 Dehydrospermidine
- C17010 Dehypoxanthine futalosine
- C21084 Demethylphylloquinol
- C00330 Deoxyguanosine
- C00677 Deoxynucleoside triphosphate
- C00882 Dephospho-CoA
- C04666 D-erythro-1-(Imidazol-4-yl)glycerol 3-phosphate
- C00279 D-Erythrose 4-phosphate
- C00354 D-Fructose 1,6-bisphosphate
- C01094 D-Fructose 1-phosphate
- C00085 D-Fructose 6-phosphate
- C00905 D-Fructuronate
- C01113 D-Galactose 6-phosphate
- C00333 D-Galacturonate
- C00257 D-Gluconic acid
- C01236 D-Glucono-1,5-lactone 6-phosphate
- C00329 D-Glucosamine
- C00352 D-Glucosamine 6-phosphate
- C00103 D-Glucose 1-phosphate
- C00092 D-Glucose 6-phosphate
- C00217 D-Glutamate
- C00118 D-Glyceraldehyde 3-phosphate
- C00258 D-Glycerate
- C19879 D-glycero-alpha-D-manno-Heptose 1,7-bisphosphate
- C19882 D-glycero-D-manno-Heptose 7-phosphate
- C02965 D-Hexose 6-phosphate
- C06419 D-Histidine
- C05925 Dihydroneopterin phosphate
- C20300 Dimethylarsinous acid
- C00013 Diphosphate
- C04574 di-trans,poly-cis-Undecaprenyl diphosphate
- C17556 di-trans,poly-cis-Undecaprenyl phosphate
- C00159 D-Mannose
- C00636 D-Mannose 1-phosphate
- C00275 D-Mannose 6-phosphate
- C04299 D-myo-Inositol 1,2-cyclic phosphate
- C00039 DNA
- C03391 DNA 6-methylaminopurine
- C01246 Dolichyl beta-D-glucosyl phosphate
- C00621 Dolichyl diphosphate
- C00515 D-Ornithine
- C01151 D-Ribose 1,5-bisphosphate
- C00117 D-Ribose 5-phosphate
- C00199 D-Ribulose 5-phosphate
- C01097 D-Tagatose 6-phosphate
- C11930 dTDP-2,6-dideoxy-D-glycero-hex-2-enos-4-ulose
- C06620 dTDP-3,4-dioxo-2,6-dideoxy-D-glucose
- C00688 dTDP-4-dehydro-beta-L-rhamnose
- C00842 dTDP-glucose
- C16581 D-threo-Aldono-1,5-lactone
- C00364 dTMP
- C00365 dUMP
- C00310 D-Xylulose
- C00231 D-Xylulose 5-phosphate
- C05359 e-
- C00016 FAD
- C00923 Ferricytochrome
- C00125 Ferricytochrome c
- C00126 Ferrocytochrome c
- C00406 Feruloyl-CoA
- C05199 Flavodoxin semiquinone
- C06108 Fluoroacetate
- C00061 FMN
- C00798 Formyl-CoA
- C00122 Fumarate
- C16999 Futalosine
- C02686 Galactosylceramide
- C00035 GDP
- C01222 GDP-4-dehydro-6-deoxy-D-mannose
- C19881 GDP-D-glycero-alpha-D-manno-heptose
- C00096 GDP-mannose
- C00341 Geranyl diphosphate
- C21217 Geranylgeranyl bacteriochlorophyllide a
- C00353 Geranylgeranyl diphosphate
- C02282 Glutaminyl-tRNA
- C00051 Glutathione
- C00127 Glutathione disulfide
- C05730 Glutathionylspermidine
- C00116 Glycerol
- C00111 Glycerone phosphate
- C00037 Glycine
- C00160 Glycolate
- C02412 Glycyl-tRNA(Gly)
- C00144 GMP
- C00581 Guanidinoacetate
- C00242 Guanine
- C01228 Guanosine 3′,5′-bis(diphosphate)
- C04494 Guanosine 3′-diphosphate 5′-triphosphate
- C19871 Guanylyl molybdenum cofactor
- C00080 H+
- C00462 Halide
- C04649 Heparan sulfate N-acetyl-alpha-D-glucosaminide
- C06250 Holo-[carboxylase]
- C00282 Hydrogen
- C01528 Hydrogen selenide
- C00283 Hydrogen sulfide
- C06399 Hydrogenobyrinate
- C06503 Hydrogenobyrinate a,c diamide
- C15603 Hydroquinone
- C00530 Hydroquinone
- C00262 Hypoxanthine
- C00130 IMP
- C00463 Indole
- C00311 Isocitrate
- C03281 Kanamycin A 3′-phosphate
- C06026 KDO2-lipid A
- C03508 L-2-Amino-3-oxobutanoic acid
- C05231 L-3-Aminobutyryl-CoA
- C00041 L-Alanine
- C20958 L-Alanyl-L-glutamate
- C00886 L-Alanyl-tRNA
- C00062 L-Arginine
- C02163 L-Arginyl-tRNA(Arg)
- C00152 L-Asparagine
- C03402 L-Asparaginyl-tRNA(Asn)
- C00049 L-Aspartate
- C00441 L-Aspartate 4-semialdehyde
- C20750 L-Carnitinyl-CoA
- C02291 L-Cystathionine
- C00506 L-Cysteate
- C00097 L-Cysteine
- C02882 L-Cysteine-S-conjugate
- C03125 L-Cysteinyl-tRNA(Cys)
- C02045 L-Erythrulose
- C00025 L-Glutamate
- C00064 L-Glutamine
- C03287 L-Glutamyl 5-phosphate
- C02987 L-Glutamyl-tRNA(Glu)
- C00135 L-Histidine
- C02988 L-Histidyl-tRNA(His)
- C00155 L-Homocysteine
- C16238 Lipoyl-AMP
- C00666 LL-2,6-Diaminoheptanedioate
- C21386 L-Leucyl-L-arginyl-protein
- C21457 L-Leucyl-L-aspartyl-protein
- C21455 L-Leucyl-L-glutamyl-protein
- C21387 L-Leucyl-L-lysyl-protein
- C02047 L-Leucyl-tRNA
- C00047 L-Lysine
- C01931 L-Lysyl-tRNA
- C00073 L-Methionine
- C15999 L-Methionine (S)-S-oxide
- C02430 L-Methionyl-tRNA
- C01826 L-Norvaline
- C02434 Long-chain ester
- C00077 L-Ornithine
- C03511 L-Phenylalanyl-tRNA(Phe)
- C02702 L-Prolyl-tRNA(Pro)
- C00508 L-Ribulose
- C01101 L-Ribulose 5-phosphate
- C02553 L-Seryl-tRNA(Ser)
- C00188 L-Threonine
- C20641 L-Threonylcarbamoyladenylate
- C02992 L-Threonyl-tRNA(Thr)
- C02839 L-Tyrosyl-tRNA(Tyr)
- C02554 L-Valyl-tRNA(Val)
- C04536 Magnesium protoporphyrin monomethyl ester
- C01209 Malonyl-[acyl-carrier protein]
- C00083 Malonyl-CoA
- C01935 Maltodextrin
- C02995 Maltose 6′-phosphate
- C04809 Membrane-derived-oligosaccharide 6-(glycerophospho)-D-glucose
- C05819 Menaquinol
- C01732 Mesaconate
- C00680 meso-2,6-Diaminoheptanedioate
- C11440 Methionyl peptide
- C00546 Methylglyoxal
- C05924 Molybdopterin
- C06717 Mycothiol
- C00137 myo-Inositol
- C15854 N-(4-Aminobutylidene)-[eIF5A-precursor]-lysine
- C04916 N-(5′-Phospho-D-1′-ribulosylformimino)-5-amino-1-(5”-phospho-D-ribosyl)-4-imidazolecarboxamide
- C03406 N-(L-Arginino)succinate
- C20562 N-[(7,8-Dihydropterin-6-yl)methyl]-4-(beta-D-ribofuranosyl)aniline 5′-phosphate
- C21009 N4-Aminopropylspermidine
- C21010 N4-Bis(aminopropyl)spermidine
- C22033 N5-(Cytidine 5′-diphosphoramidyl)-L-glutamine
- C03794 N6-(1,2-Dicarboxyethyl)-AMP
- C20751 N6-L-Threonylcarbamoyladenine in tRNA
- C04501 N-Acetyl-alpha-D-glucosamine 1-phosphate
- C20390 N-Acetyl-alpha-D-glucosaminyl-diphospho-trans,octacis-decaprenol
- C00140 N-Acetyl-D-glucosamine
- C00357 N-Acetyl-D-glucosamine 6-phosphate
- C01289 N-Acetyl-D-glucosaminyldiphosphoundecaprenol
- C02297 N-Acetyldiamine
- C00645 N-Acetyl-D-mannosamine
- C05539 N-Acetyl-L-2-amino-6-oxopimelate
- C00624 N-Acetyl-L-glutamate
- C04133 N-Acetyl-L-glutamate 5-phosphate
- C01250 N-Acetyl-L-glutamate 5-semialdehyde
- C21027 N-Acetylmuramic acid alpha-1-phosphate
- C00625 N-Acyl-D-mannosamine
- C00003 NAD+
- C00004 NADH
- C00006 NADP+
- C00005 NADPH
- C00438 N-Carbamoyl-L-aspartate
- C00454 NDP
- C19609 Nickel(2+)
- C00153 Nicotinamide
- C00455 Nicotinamide D-ribonucleotide
- C00253 Nicotinate
- C00533 Nitric oxide
- C02565 N-Methylhydantoin
- C21113 NMNH
- C03523 N-Substituted amino acid
- C04462 N-Succinyl-2-L-amino-6-oxoheptanedioate
- C03419 Nucleoside 3′-phosphate
- C01117 Nucleoside 5′-phosphate
- C01077 O-Acetyl-L-homoserine
- C00979 O-Acetyl-L-serine
- C04260 O-D-Alanyl-poly(ribitol phosphate)
- C01102 O-Phospho-L-homoserine
- C00295 Orotate
- C00009 Orthophosphate
- C01118 O-Succinyl-L-homoserine
- C00313 Oxalyl-CoA
- C00139 Oxidized ferredoxin
- C02869 Oxidized flavodoxin
- C00007 Oxygen
- C01134 Pantetheine 4′-phosphate
- C00864 Pantothenate
- C00472 p-Benzoquinone
- C06564 Penicillin N
- C15653 Peptide-L-methionine (R)-S-oxide
- C03895 Peptide-L-methionine (S)-S-oxide
- C03633 Peptidylproline (omega=0)
- C15584 Phenol
- C00582 Phenylacetyl-CoA
- C00166 Phenylpyruvate
- C00416 Phosphatidate
- C00344 Phosphatidylglycerol
- C04308 Phosphatidyl-N-dimethylethanolamine
- C01241 Phosphatidyl-N-methylethanolamine
- C00074 Phosphoenolpyruvate
- C03167 Phosphonoacetaldehyde
- C00562 Phosphoprotein
- C02741 Phosphoribosyl-AMP
- C03313 Phylloquinol
- C16695 Plastoquinol-9
- C00404 Polyphosphate
- C19692 Polysulfide
- C06407 Precorrin 4
- C06416 Precorrin 5
- C06408 Precorrin 8X
- C00254 Prephenate
- C21177 Prokaryotic ubiquitin-like protein
- C00615 Protein histidine
- C02188 Protein lysine
- C04261 Protein N(pi)-phospho-L-histidine
- C16237 Protein N6-(lipoyl)lysine
- C20743 Protein N6-acetyl-L-lysine
- C00585 Protein tyrosine
- C02880 Protochlorophyllide
- C20082 Pseudaminic acid
- C01168 Pseudouridine 5′-phosphate
- C21178 Pupylated protein
- C15587 Purine
- C21851 Pyridinium-3,5-biscarboxylic acid mononucleotide
- C21767 Pyridinium-3,5-bisthiocarboxylate mononucleotide
- C21878 Pyridinium-3-carboxy-5-thiocarboxylic acid mononucleotide
- C00250 Pyridoxal
- C00627 Pyridoxine phosphate
- C00022 Pyruvate
- C22150 Reduced [2Fe-2S] ferredoxin
- C01080 Reduced coenzyme F420
- C02745 Reduced flavodoxin
- C22336 Reduced hydrogenase
- C00376 Retinal
- C00046 RNA
- C04425 S-Adenosyl-4-methylthio-2-oxobutanoate
- C00021 S-Adenosyl-L-homocysteine
- C00019 S-Adenosyl-L-methionine
- C00805 Salicylate
- C05382 Sedoheptulose 7-phosphate
- C06718 S-Formylmycothiol
- C03431 S-Inosyl-L-homocysteine
- C05778 Sirohydrochlorin
- C22360 S-methyl-1-thio-D-ribulose 5-phosphate
- C22359 S-Methyl-1-thio-D-xylulose 5-phosphate
- C00623 sn-Glycerol 1-phosphate
- C00093 sn-Glycerol 3-phosphate
- C08250 Sophorose
- C03539 S-Ribosyl-L-homocysteine
- C05824 S-Sulfo-L-cysteine
- C03641 Sterol 3-beta-D-glucoside
- C01138 Streptomycin 6-phosphate
- C00059 Sulfate
- C02084 Tetrathionate
- C03541 THF-polyglutamate
- C01081 Thiamin monophosphate
- C00378 Thiamine
- C15814 Thiocarboxy-[sulfur-carrier protein]
- C01755 Thiocyanate
- C00145 Thiol
- C00343 Thioredoxin disulfide
- C20369 trans,octacis-Decaprenylphospho-beta-D-erythro-pentofuranosid-2-ulose
- C00448 trans,trans-Farnesyl diphosphate
- C00422 Triacylglycerol
- C00066 tRNA
- C04728 tRNA containing 5-[(methylamino)methyl]-2-thiouridylate
- C19080 tRNA with a 3′ CC end
- C19085 tRNA with a 3′ CCA end
- C01635 tRNA(Ala)
- C01642 tRNA(Gly)
- C01645 tRNA(Leu)
- C00390 Ubiquinol
- C00015 UDP
- C19725 UDP-2,3-diacetamido-2,3-dideoxy-alpha-D-glucuronate
- C19745 UDP-2,3-diacetamido-2,3-dideoxy-alpha-D-mannuronate
- C19823 UDP-2-acetamido-2,6-dideoxy-beta-L-arabino-hexos-4-ulose
- C20395 UDP-2-acetamido-2-deoxy-alpha-D-ribo-hex-3-uluronate
- C20849 UDP-2-acetamido-3-dehydro-2-deoxy-alpha-D-glucopyranose
- C04613 UDP-2-acetamido-4-dehydro-2,6-dideoxyglucose
- C03733 UDP-alpha-D-galactofuranose
- C00052 UDP-alpha-D-galactose
- C00190 UDP-D-xylose
- C00167 UDP-glucuronate
- C04702 UDPMurNAc(oyl-L-Ala-D-gamma-Glu-L-Lys-D-Ala-D-Ala)
- C20357 UDP-N,N’-diacetylbacillosamine
- C04631 UDP-N-acetyl-3-(1-carboxyvinyl)-D-glucosamine
- C00043 UDP-N-acetyl-alpha-D-glucosamine
- C22053 UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-L-glutamate
- C00203 UDP-N-acetyl-D-galactosamine
- C01212 UDP-N-acetylmuramoyl-L-alanine
- C00692 UDP-N-acetylmuramoyl-L-alanyl-D-glutamate
- C00105 UMP
- C00106 Uracil
- C05326 Uridylyl-[protein-PII]
- C00001 Water
- C00385 Xanthine
- C00655 Xanthosine 5′-phosphate
For Metabolite Substrate/Consumers
- C05366 (+)-Pinoresinol
- C04411 (2R,3S)-3-Isopropylmalate
- C06326 (2S)-2-{[1-(R)-Carboxyethyl]amino}pentanoate
- C04236 (2S)-2-Isopropyl-3-oxosuccinate
- C04593 (2S,3R)-3-Hydroxybutane-1,2,3-tricarboxylate
- C01142 (3S)-3,6-Diaminohexanoate
- C01186 (3S,5S)-3,5-Diaminohexanoate
- C05786 (3Z)-Phycocyanobilin
- C20485 (4S)-4-Hydroxy-2-oxoglutarate
- C20954 (5-Formylfuran-3-yl)methyl phosphate
- C04856 (6S)-6beta-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- C04899 (6S)-6beta-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide phosphate
- C02214 (E)-Glutaconate
- C22385 (L-Cysteinyl)adenylate
- C03195 (R)-10-Hydroxystearate
- C04272 (R)-2,3-Dihydroxy-3-methylbutanoate
- C01087 (R)-2-Hydroxyglutarate
- C02612 (R)-2-Methylmalate
- C04352 (R)-4′-Phosphopantothenoyl-L-cysteine
- C00810 (R)-Acetoin
- C16241 (R)-Lipoate
- C02912 (R)-Propane-1,2-diol
- C03044 (R,R)-Butane-2,3-diol
- C00898 (R,R)-Tartaric acid
- C02103 (S)-2-Haloacid
- C15565 (S)-2-Hydroxyacid
- C11499 (S)-3-Sulfolactate
- C00337 (S)-Dihydroorotate
- C02917 (S)-Propane-1,2-diol
- C07281 [eIF5A-precursor]-lysine
- C15811 [Enzyme]-cysteine
- C15812 [Enzyme]-S-sulfanylcysteine
- C22154 [Fe-S] cluster scaffold protein carrying a [4Fe-4S]2+ cluster
- C01281 [L-Glutamate:ammonia ligase (ADP-forming)]
- C16240 [Lipoyl-carrier protein]-L-lysine
- C21179 [Prokaryotic ubiquitin-like protein]-L-glutamine
- C02743 [Protein]-L-cysteine
- C19803 [Protein]-L-threonine
- C16236 [Protein]-N6-(octanoyl)-L-lysine
- C01242 [Protein]-S8-aminomethyldihydrolipoyllysine
- C21440 [Protein]-S-sulfanyl-L-cysteine
- C05250 [Protein-PII]
- C21879 1-(5-O-Phospho-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium-3-carbonyl adenylate
- C04751 1-(5-Phospho-D-ribosyl)-5-amino-4-imidazolecarboxylate
- C02739 1-(5-Phospho-D-ribosyl)-ATP
- C04823 1-(5′-Phosphoribosyl)-5-amino-4-(N-succinocarboxamide)-imidazole
- C04677 1-(5′-Phosphoribosyl)-5-amino-4-imidazolecarboxamide
- C02490 1,2-beta-D-Glucan
- C21335 1,2-beta-Oligomannan
- C06364 1,2-Diacyl-3-alpha-D-glucosyl-sn-glycerol
- C00641 1,2-Diacyl-sn-glycerol
- C19776 1,2-Diacyl-sn-glycerol 3-diphosphate
- C17207 1,4-beta-D-Mannooligosaccharide
- C16538 1,5-Anhydro-D-mannitol
- C19769 1,6-Anhydro-N-acetyl-beta-muramate
- C00234 10-Formyltetrahydrofolate
- C00681 1-Acyl-sn-glycerol 3-phosphate
- C01234 1-Aminocyclopropane-1-carboxylate
- C11811 1-Hydroxy-2-methyl-2-butenyl 4-diphosphate
- C19703 1-O-[2-(L-Cysteinamido)-2-deoxy-alpha-D-glucopyranosyl]-1D-myo-inositol
- C01194 1-Phosphatidyl-D-myo-inositol
- C04637 1-Phosphatidyl-D-myo-inositol 4,5-bisphosphate
- C20247 2-(2-Carboxy-4-methylthiazol-5-yl)ethyl phosphate
- C19771 2′-(5-Triphosphoribosyl)-3′-dephospho-CoA
- C04640 2-(Formamido)-N1-(5′-phosphoribosyl)acetamidine
- C03972 2,3,4,5-Tetrahydrodipicolinate
- C01159 2,3-Bisphospho-D-glycerate
- C01240 2′,3′-Cyclic nucleotide
- C04039 2,3-Dihydroxy-3-methylbutanoate
- C20246 2-[(2R,5Z)-2-Carboxy-4-methylthiazol-5(2H)-ylidene]ethyl phosphate
- C21895 2-Acetylphloroglucinol
- C17234 2-Aminobut-2-enoate
- C22039 2-Carboxy-1,4-naphthoquinone
- C11434 2-C-Methyl-D-erythritol 4-phosphate
- C04442 2-Dehydro-3-deoxy-6-phospho-D-gluconate
- C04691 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate
- C03826 2-Dehydro-3-deoxy-D-xylonate
- C19969 2-Hydroxy-dATP
- C05123 2-Hydroxyethanesulfonate
- C06451 2-Hydroxyethylphosphonate
- C20905 2-Iminobutanoate
- C20904 2-Iminopropanoate
- C02631 2-Isopropylmaleate
- C19858 2-Methoxy-6-all-trans-polyprenyl-1,4-benzoquinol
- C15882 2-Methyl-6-phytylquinol
- C17570 2-Methyl-6-solanyl-1,4-benzoquinol
- C21609 2-O-(6-O-Acyl-alpha-D-mannosyl)-1-phosphatidyl-1D-myo-inositol
- C19791 2-O-(alpha-D-Glucopyranosyl)-3-phospho-D-glycerate
- C00026 2-Oxoglutarate
- C11436 2-Phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- C00631 2-Phospho-D-glycerate
- C00988 2-Phosphoglycolate
- C13309 2-Phytyl-1,4-naphthoquinone
- C20772 3-[(1-Carboxyvinyl)oxy]benzoate
- C11638 3-Amino-2-oxopropyl phosphate
- C04287 3D-3,5/4-Trihydroxycyclohexane-1,2-dione
- C00944 3-Dehydroquinate
- C21383 3-Deoxy-D-glycero-D-galacto-non-2-ulopyranosonate 9-phosphate
- C02514 3-Fumarylpyruvate
- C00141 3-Methyl-2-oxobutanoic acid
- C02939 3-Methylbutanoyl-CoA
- C22131 3”-O-Acetyl-ADP-D-ribose
- C02940 3-Oxo-5alpha-steroid
- C01876 3-Oxosteroid
- C00197 3-Phospho-D-glycerate
- C02798 3-Phosphonopyruvate
- C02508 3′-Ribonucleotide
- C22312 3-Sulfanylpropanoate
- C06118 4-(4-Deoxy-alpha-D-gluc-4-enuronosyl)-D-galacturonate
- C20559 4-(beta-D-Ribofuranosyl)aniline 5′-phosphate
- C11435 4-(Cytidine 5′-diphospho)-2-C-methyl-D-erythritol
- C04556 4-Amino-2-methyl-5-(phosphooxymethyl)pyrimidine
- C20267 4-Amino-5-aminomethyl-2-methylpyrimidine
- C01279 4-Amino-5-hydroxymethyl-2-methylpyrimidine
- C04752 4-Amino-5-hydroxymethyl-2-methylpyrimidine diphosphate
- C05848 4-Hydroxy-3-polyprenylbenzoate
- C00989 4-Hydroxybutanoic acid
- C13635 4-Hydroxyphenacyl alcohol
- C04327 4-Methyl-5-(2-phosphooxyethyl)thiazole
- C20236 4-O-beta-D-Mannopyranosyl-D-glucopyranose
- C20749 4-Trimethylammoniobutanoyl-CoA
- C04294 5-(2-Hydroxyethyl)-4-methylthiazole
- C04896 5-(5-Phospho-D-ribosylaminoformimino)-1-(5-phosphoribosyl)-imidazole-4-carboxamide
- C00445 5,10-Methenyltetrahydrofolate
- C00143 5,10-Methylenetetrahydrofolate
- C04454 5-Amino-6-(5′-phospho-D-ribitylamino)uracil
- C00990 5-Aminopentanamide
- C21877 5-Carboxy-1-(5-O-phospho-beta-D-ribofuranosyl)pyridin-1-ium-3-carbonyl adenylate
- C15667 5-Carboxyamino-1-(5-phospho-D-ribosyl)imidazole
- C05198 5′-Deoxyadenosine
- C16737 5-Deoxy-D-glucuronate
- C00664 5-Formiminotetrahydrofolate
- C00170 5′-Methylthioadenosine
- C01269 5-O-(1-Carboxyvinyl)-3-phosphoshikimate
- C01879 5-Oxoproline
- C03090 5-Phosphoribosylamine
- C04376 5′-Phosphoribosyl-N-formylglycinamide
- C02520 5′-Ribonucleotide
- C20773 6-Amino-6-deoxyfutalosine
- C20737 6-Geranylgeranyl-2-methylbenzene-1,4-diol
- C03847 6-Phospho-beta-D-galactoside
- C15858 7,9,7′,9′-tetracis-Lycopene
- C19759 7,9,9′-tricis-Neurosporene
- C01092 8-Amino-7-oxononanoate
- C19967 8-Oxo-dGTP
- C22234 8-Oxo-GTP
- C15857 9,9′-dicis-zeta-Carotene
- C00033 Acetate
- C00024 Acetyl-CoA
- C00173 Acyl-[acyl-carrier protein]
- C00040 Acyl-CoA
- C06506 Adenosyl cobyrinate a,c diamide
- C01299 Adenylyl-[L-glutamate:ammonia ligase (ADP-forming)]
- C15813 Adenylyl-[sulfur-carrier protein]
- C00008 ADP
- C00498 ADP-glucose
- C00301 ADP-ribose
- C03687 Alkane-alpha,omega-diamine
- C05847 all-trans-Polyprenyl diphosphate
- C04218 alpha,alpha’-Trehalose 6-mycolate
- C00984 alpha-D-Galactose
- C00446 alpha-D-Galactose 1-phosphate
- C06156 alpha-D-Glucosamine 1-phosphate
- C00267 alpha-D-Glucose
- C00668 alpha-D-Glucose 6-phosphate
- C20440 alpha-D-Ribose 1,2-cyclic phosphate 5-phosphate
- C00620 alpha-D-Ribose 1-phosphate
- C20237 alpha-Maltose 1-phosphate
- C03373 Aminoimidazole ribotide
- C00014 Ammonia
- C00020 AMP
- C20144 Androsta-1,4-diene-3,17-dione
- C06249 Apo-[carboxylase]
- C11215 Arsenate ion
- C00002 ATP
- C11242 Bacteriochlorophyll a
- C21336 beta-1,2-Mannobiose
- C21454 beta-1,2-Mannotriose
- C00099 beta-Alanine
- C00602 beta-D-Galactoside
- C00663 beta-D-Glucose 1-phosphate
- C20887 beta-D-Mannosyl-1,4-N-acetyl-D-glucosamine
- C20568 beta-L-Arabinofuranosyl-(1->2)-beta-L-arabinofuranose
- C00486 Bilirubin
- C00120 Biotin
- C00323 Caffeoyl-CoA
- C01563 Carbamate
- C00169 Carbamoyl phosphate
- C01353 Carbonic acid
- C04419 Carboxybiotin-carboxyl-carrier protein
- C20969 Carboxyphosphate
- C00513 CDP-glycerol
- C00185 Cellobiose
- C00916 Cephalosporin C
- C00695 Cholic acid
- C00588 Choline phosphate
- C00919 Choline sulfate
- C04431 cis-4-Carboxymethylenebut-2-en-4-olide
- C00417 cis-Aconitate
- C00158 Citrate
- C00128 CMP-N-acetylneuraminate
- C00011 CO2
- C17401 Cobalt-factor III
- C11540 Cobalt-precorrin 4
- C11545 Cobalt-precorrin 8
- C11538 Cobalt-sirohydrochlorin
- C05773 Cobyrinate
- C19724 Cobyrinate c-monamide
- C04628 Coenzyme B
- C00876 Coenzyme F420
- C03576 Coenzyme M
- C00791 Creatinine
- C00063 CTP
- C00177 Cyanide ion
- C00973 Cyclomaltodextrin
- C00475 Cytidine
- C00380 Cytosine
- C03492 D-4′-Phosphopantothenate
- C00239 dCMP
- C00857 Deamino-NAD+
- C02970 Decaprenol phosphate
- C15494 Decylubiquinone
- C15853 Dehydrospermidine
- C17010 Dehypoxanthine futalosine
- C19847 Demethylmenaquinol
- C21084 Demethylphylloquinol
- C00677 Deoxynucleoside triphosphate
- C00882 Dephospho-CoA
- C04666 D-erythro-1-(Imidazol-4-yl)glycerol 3-phosphate
- C00279 D-Erythrose 4-phosphate
- C00095 D-Fructose
- C00354 D-Fructose 1,6-bisphosphate
- C00085 D-Fructose 6-phosphate
- C01113 D-Galactose 6-phosphate
- C00257 D-Gluconic acid
- C00198 D-Glucono-1,5-lactone
- C01236 D-Glucono-1,5-lactone 6-phosphate
- C00031 D-Glucose
- C00103 D-Glucose 1-phosphate
- C00217 D-Glutamate
- C00118 D-Glyceraldehyde 3-phosphate
- C00258 D-Glycerate
- C19880 D-glycero-alpha-D-manno-Heptose 1-phosphate
- C19878 D-glycero-alpha-D-manno-Heptose 7-phosphate
- C00286 dGTP
- C00738 D-Hexose
- C05196 Dihydroflavodoxin
- C00921 Dihydropteroate
- C20522 Dihydrourocanate
- C00235 Dimethylallyl diphosphate
- C00013 Diphosphate
- C01345 dITP
- C04574 di-trans,poly-cis-Undecaprenyl diphosphate
- C17556 di-trans,poly-cis-Undecaprenyl phosphate
- C03189 DL-Glycerol 1-phosphate
- C00476 D-Lyxose
- C00159 D-Mannose
- C00636 D-Mannose 1-phosphate
- C00039 DNA
- C00821 DNA adenine
- C04213 Dolichyl diphosphooligosaccharide
- C00515 D-Ornithine
- C01068 D-Ribitol 5-phosphate
- C00121 D-Ribose
- C00117 D-Ribose 5-phosphate
- C00309 D-Ribulose
- C00199 D-Ribulose 5-phosphate
- C11930 dTDP-2,6-dideoxy-D-glycero-hex-2-enos-4-ulose
- C04346 dTDP-4-amino-4,6-dideoxy-D-galactose
- C11922 dTDP-4-oxo-2,6-dideoxy-D-glucose
- C11907 dTDP-4-oxo-6-deoxy-D-glucose
- C00842 dTDP-glucose
- C03319 dTDP-L-rhamnose
- C02143 D-threo-Aldose
- C00459 dTTP
- C00365 dUMP
- C00460 dUTP
- C00231 D-Xylulose 5-phosphate
- C05359 e-
- C15973 Enzyme N6-(dihydrolipoyl)lysine
- C02391 Ester
- C00189 Ethanolamine
- C00016 FAD
- C00125 Ferricytochrome c
- C00924 Ferrocytochrome
- C00126 Ferrocytochrome c
- C19775 Fluoroacetyl-CoA
- C00061 FMN
- C03479 Folinic acid
- C00067 Formaldehyde
- C00488 Formamide
- C00058 Formate
- C00798 Formyl-CoA
- C11439 Formyl-L-methionyl peptide
- C16999 Futalosine
- C00325 GDP-L-fucose
- C00096 GDP-mannose
- C00341 Geranyl diphosphate
- C07292 Glutaredoxin
- C00051 Glutathione
- C05730 Glutathionylspermidine
- C00116 Glycerol
- C00184 Glycerone
- C03120 Glycerophosphodiester
- C02412 Glycyl-tRNA(Gly)
- C00144 GMP
- C00044 GTP
- C01228 Guanosine 3′,5′-bis(diphosphate)
- C00080 H+
- C00288 HCO3-
- C04384 Heparan sulfate alpha-D-glucosaminide
- C00205 hn
- C06250 Holo-[carboxylase]
- C06399 Hydrogenobyrinate
- C00530 Hydroquinone
- C00168 Hydroxypyruvate
- C22226 Icosanoyl-[(phenol)carboxyphthiodiolenone synthase]
- C00130 IMP
- C00331 Indolepyruvate
- C00885 Isochorismate
- C00311 Isocitrate
- C17658 Isolithocholate
- C05557 Isopenicillin N
- C00129 Isopentenyl diphosphate
- C00081 ITP
- C00304 Kanamycin
- C03508 L-2-Amino-3-oxobutanoic acid
- C00041 L-Alanine
- C20957 L-Alanyl-D-glutamate
- C20925 L-Alanyl-gamma-D-glutamyl-meso-2,6-diaminoheptanedioate
- C05519 L-Allothreonine
- C00259 L-Arabinose
- C16739 L-Arginyl-protein
- C00152 L-Asparagine
- C00049 L-Aspartate
- C00441 L-Aspartate 4-semialdehyde
- C21458 L-Aspartyl-protein
- C06251 Lauroyl-KDO2-lipid IV(A)
- C00318 L-Carnitine
- C00327 L-Citrulline
- C00097 L-Cysteine
- C02882 L-Cysteine-S-conjugate
- C01720 L-Fuconate
- C00025 L-Glutamate
- C01165 L-Glutamate 5-semialdehyde
- C00064 L-Glutamine
- C02986 L-Glutaminyl-peptide
- C21456 L-Glutamyl-protein
- C00860 L-Histidinol
- C01100 L-Histidinol phosphate
- C00263 L-Homoserine
- C00770 L-Idonate
- C16238 Lipoyl-AMP
- C02051 Lipoylprotein
- C00666 LL-2,6-Diaminoheptanedioate
- C00123 L-Leucine
- C02047 L-Leucyl-tRNA
- C00047 L-Lysine
- C21388 L-Lysyl-protein
- C00073 L-Methionine
- C00638 Long-chain fatty acid
- C00077 L-Ornithine
- C00079 L-Phenylalanine
- C00148 L-Proline
- C00508 L-Ribulose
- C05688 L-Selenocysteine
- C00065 L-Serine
- C00188 L-Threonine
- C12147 L-Threonine O-3-phosphate
- C20641 L-Threonylcarbamoyladenylate
- C00183 L-Valine
- C03516 Magnesium protoporphyrin
- C00083 Malonyl-CoA
- C04566 Membrane-derived-oligosaccharide D-glucose
- C00409 Methanethiol
- C17530 Methyl acetate
- C03620 Monocarboxylic acid amide
- C15585 myo-Inositol phosphate
- C15854 N-(4-Aminobutylidene)-[eIF5A-precursor]-lysine
- C04916 N-(5′-Phospho-D-1′-ribulosylformimino)-5-amino-1-(5”-phospho-D-ribosyl)-4-imidazolecarboxamide
- C03406 N-(L-Arginino)succinate
- C20418 N,N’-Diacetyllegionaminate
- C22293 N4-Acetylcytidine
- C21009 N4-Aminopropylspermidine
- C21785 N5-Phospho-L-glutamine
- C03794 N6-(1,2-Dicarboxyethyl)-AMP
- C04501 N-Acetyl-alpha-D-glucosamine 1-phosphate
- C00140 N-Acetyl-D-glucosamine
- C00357 N-Acetyl-D-glucosamine 6-phosphate
- C00645 N-Acetyl-D-mannosamine
- C00624 N-Acetyl-L-glutamate
- C01250 N-Acetyl-L-glutamate 5-semialdehyde
- C03708 N-Acetyl-O-acetylneuraminate
- C00437 N-Acetylornithine
- C03000 N-Acyl-D-glucosamine
- C00625 N-Acyl-D-mannosamine
- C00003 NAD+
- C00004 NADH
- C00006 NADP+
- C00005 NADPH
- C15586 N-D-Ribosylpurine
- C21769 Ni(II)-pyridinium-3,5-bisthiocarboxylate mononucleotide
- C00455 Nicotinamide D-ribonucleotide
- C00244 Nitrate
- C00533 Nitric oxide
- C00697 Nitrogen
- C00887 Nitrous oxide
- C03880 N-Substituted aminoacyl-tRNA
- C04421 N-Succinyl-LL-2,6-diaminoheptanedioate
- C01075 N-Sulfo-D-glucosamine
- C21158 Nucleoside 3′,5′-bisphosphate
- C00201 Nucleoside triphosphate
- C01077 O-Acetyl-L-homoserine
- C00979 O-Acetyl-L-serine
- C05752 Octanoyl-[acp]
- C06055 O-Phospho-4-hydroxy-L-threonine
- C01102 O-Phospho-L-homoserine
- C01005 O-Phospho-L-serine
- C01103 Orotidine 5′-phosphate
- C00009 Orthophosphate
- C01118 O-Succinyl-L-homoserine
- C00209 Oxalate
- C00036 Oxaloacetate
- C00313 Oxalyl-CoA
- C22151 Oxidized [2Fe-2S] ferredoxin
- C02869 Oxidized flavodoxin
- C22335 Oxidized hydrogenase
- C01134 Pantetheine 4′-phosphate
- C00864 Pantothenate
- C00472 p-Benzoquinone
- C00395 Penicillin
- C03798 Peptidylproline (omega=180)
- C07086 Phenylacetic acid
- C00416 Phosphatidate
- C00350 Phosphatidylethanolamine
- C00344 Phosphatidylglycerol
- C03892 Phosphatidylglycerophosphate
- C01241 Phosphatidyl-N-methylethanolamine
- C00074 Phosphoenolpyruvate
- C02741 Phosphoribosyl-AMP
- C05427 Phytyl diphosphate
- C00653 Poly(ribitol phosphate)
- C00404 Polyphosphate
- C19692 Polysulfide
- C02463 Precorrin 2
- C06406 Precorrin 3B
- C06407 Precorrin 4
- C06319 Precorrin 6Y
- C06408 Precorrin 8X
- C18239 Precursor Z
- C00254 Prephenate
- C03428 Presqualene diphosphate
- C21177 Prokaryotic ubiquitin-like protein
- C01845 Propan-2-ol
- C00017 Protein
- C00615 Protein histidine
- C02188 Protein lysine
- C04261 Protein N(pi)-phospho-L-histidine
- C20743 Protein N6-acetyl-L-lysine
- C01167 Protein tyrosine phosphate
- C02880 Protochlorophyllide
- C02067 Pseudouridine
- C21851 Pyridinium-3,5-biscarboxylic acid mononucleotide
- C21878 Pyridinium-3-carboxy-5-thiocarboxylic acid mononucleotide
- C00250 Pyridoxal
- C00647 Pyridoxamine phosphate
- C00314 Pyridoxine
- C00627 Pyridoxine phosphate
- C00022 Pyruvate
- C15602 Quinone
- C00030 Reduced acceptor
- C01080 Reduced coenzyme F420
- C00138 Reduced ferredoxin
- C01847 Reduced FMN
- C00473 Retinol
- C00255 Riboflavin
- C03802 Ribonucleoside triphosphate
- C00046 RNA
- C05729 R-S-Cysteinylglycine
- C18235 S-(Hydroxymethyl)mycothiol
- C00019 S-Adenosyl-L-methionine
- C06153 scyllo-Inositol
- C00447 Sedoheptulose 1,7-bisphosphate
- C05382 Sedoheptulose 7-phosphate
- C00493 Shikimate
- C00315 Spermidine
- C03539 S-Ribosyl-L-homocysteine
- C00370 Sterol
- C00232 Succinate semialdehyde
- C00089 Sucrose
- C06125 Sulfatide
- C20829 Sulfoquinovose
- C05761 Tetradecanoyl-[acp]
- C03541 THF-polyglutamate
- C00378 Thiamine
- C00342 Thioredoxin
- C00214 Thymidine
- C20370 trans,octacis-Decaprenylphospho-beta-D-arabinofuranose
- C20368 trans,octacis-Decaprenylphospho-beta-D-ribofuranose
- C20420 tritrans,heptacis-Undecaprenyl phosphate
- C17324 tRNA adenine
- C11478 tRNA containing 5-(aminomethyl)-2-thiouridine
- C02211 tRNA precursor
- C00868 tRNA uridine
- C19080 tRNA with a 3′ CC end
- C19078 tRNA with a 3′ cytidine
- C01635 tRNA(Ala)
- C01636 tRNA(Arg)
- C01639 tRNA(Cys)
- C01641 tRNA(Glu)
- C01642 tRNA(Gly)
- C01643 tRNA(His)
- C01645 tRNA(Leu)
- C01646 tRNA(Lys)
- C01647 tRNA(Met)
- C01648 tRNA(Phe)
- C01649 tRNA(Pro)
- C01650 tRNA(Ser)
- C01651 tRNA(Thr)
- C01652 tRNA(Trp)
- C00787 tRNA(Tyr)
- C01653 tRNA(Val)
- C00399 Ubiquinone
- C19725 UDP-2,3-diacetamido-2,3-dideoxy-alpha-D-glucuronate
- C20359 UDP-2-acetamido-3-amino-2,3-dideoxy-alpha-D-glucuronate
- C04630 UDP-2-acetamido-4-amino-2,4,6-trideoxy-alpha-D-glucose
- C19961 UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine
- C00052 UDP-alpha-D-galactose
- C00029 UDP-glucose
- C00167 UDP-glucuronate
- C20357 UDP-N,N’-diacetylbacillosamine
- C04573 UDP-N-acetyl-2-amino-2-deoxy-D-glucuronate
- C00043 UDP-N-acetyl-alpha-D-glucosamine
- C22053 UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-L-glutamate
- C00203 UDP-N-acetyl-D-galactosamine
- C01050 UDP-N-acetylmuramate
- C01212 UDP-N-acetylmuramoyl-L-alanine
- C00692 UDP-N-acetylmuramoyl-L-alanyl-D-glutamate
- C05892 UDP-N-acetylmuramoyl-L-alanyl-gamma-D-glutamyl-L-lysine
- C00105 UMP
- C01968 Undecaprenol
- C05893 Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl-D-alanine
- C00086 Urea
- C01051 Uroporphyrinogen III
- C00075 UTP
- C00001 Water
- C00655 Xanthosine 5′-phosphate
- C00700 XTP
- C00379 Xylitol
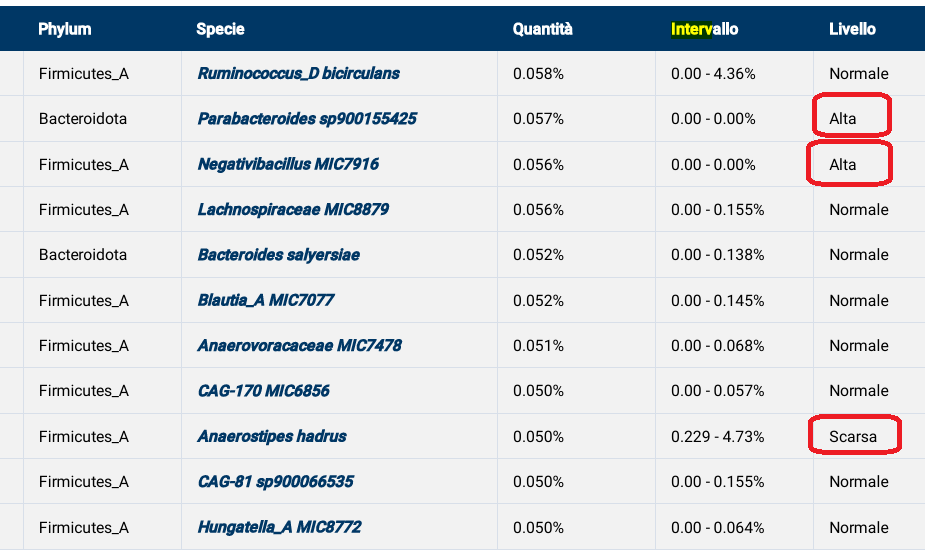


Recent Comments