A few weeks ago, I stumbled on some algorithms that had good results for predicting symptoms from bacteria. The next logical step was using the associations to get suggestions. While working on a blog post, I was getting odd results and digging into why, I discovered both logic and computational errors. Two readers had also raised questions about apparent bizarre logic. They were right — my logic was too simplistic and needed revision as well as better exposure of the logic being used.
The corrected version is up now. The main differences are:
- Auto checking check boxes will happen less
- Possible additions (unchecked) are marked with a 💡
- For Premium users, you get to see more of the gears that are turning.
To explain the issue, let us look at some details shown when in professional mode.
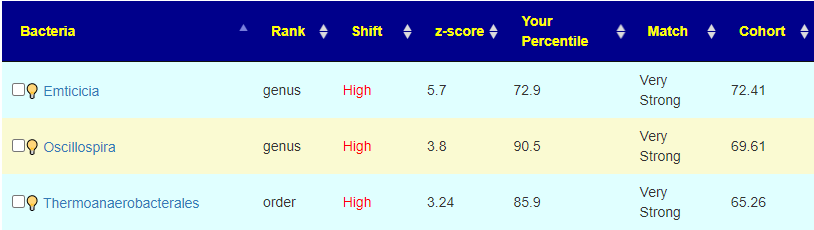
The old logic made suggestions to move away from the Cohort number. So for Emticicia, because we were higher than the Cohort value, we would try to raise it. This revision tries to lower it towards the 50%ile instead. The conceptual logic of moving away from the cohort was correct ignoring the sample percentile was incorrect. This implementation revision should correct this. For the other two bacteria above, we see that the cohort was high and the shift was even higher — again, moving away from the cohort is the desired, but moving higher than 90%ile is likely a poor choice, in this case you want to really move it down a lot.
The other factor is taking into account the z-score, etc. Some pages may have no automatic check. If you just click thru, you may get this message:
Let us look at some of the automatic checked
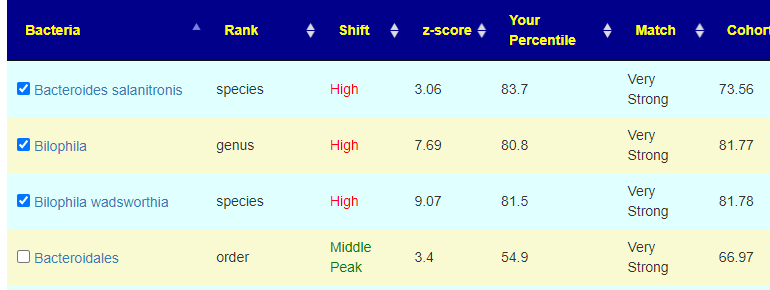
Both the person’s percentile and the Cohort are high, one was below the cohort and one was above. Because they are both high, the logic is to move them down to the middle (ignoring which side of the cohort it was on). The last one was not checked despite being Very Strong because the sample percentile was so close to the 50%ile (middle value)
A third set of examples is below, which include the weight being visible (likely will be moved to a professional feature — mainly because it can be more complicated to interpret well)
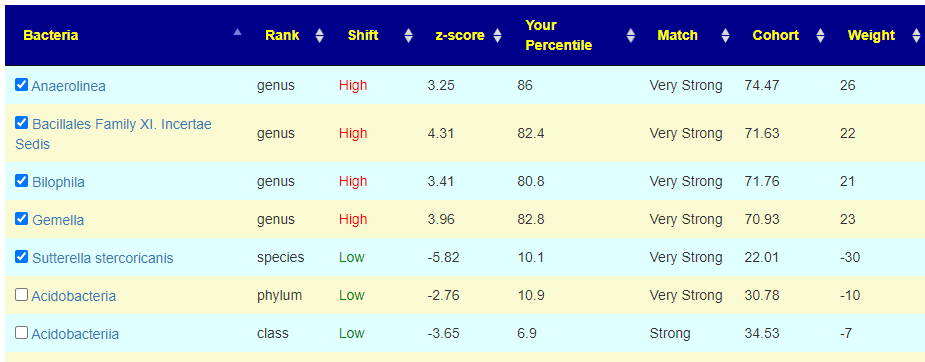
To get an automatic check the weight needs to be at least 20, for the 💡 , at least 10. For Acidobacteria, it is low but it is also a considerable distance from the cohort average. If selected, we declare a negative value and thus attempt to increase it (potentially moving it much closer to the cohort typical value — i.e. increase symptom). On the flip side, at only the 10%ile, you do not really want to decrease it more. A dilemma – excluding it is actually the best path. It has significance for the symptom forecast but has no clear action for altering the associated bacteria.
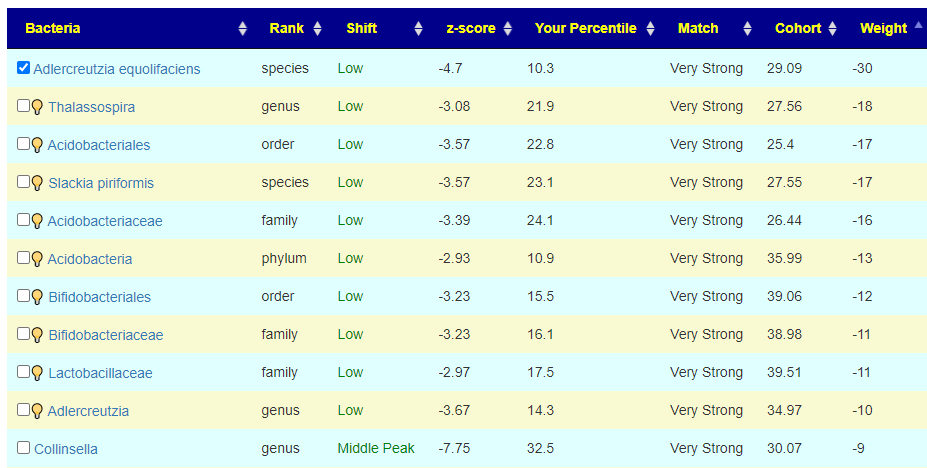
For this last one, we see Collinsella is a middle peak, and the desired direction is to increase it (negative value). Remember that these weights are used in computing the weights for suggestions.
Bottom lIne
The site is always in a state of change — from new studies being added, new samples being uploaded (and many statistics recalculated daily) and tuning and adjust algorithms — in this case readers questions lead to looking at the working data and seeing potential issues to correct (as well as displaying those numbers so people may ask questions — leading to still better algorithms).
1 thought on “Suggestions from Symptoms are Changed”
Comments are closed.